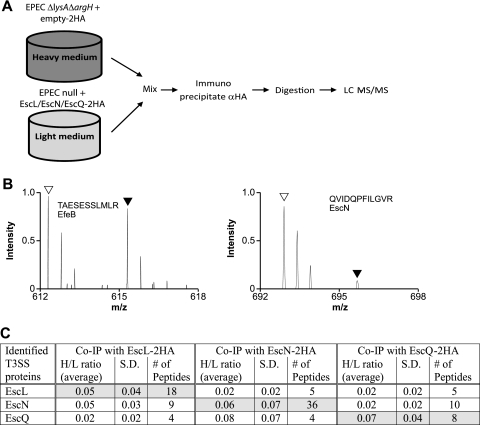
Fig. 2.
SILAC screen for interacting proteins EscL, EscN, and EscQ. (A) Schematic representation of the SILAC method. The EPECΔlysAΔargH strain expressing the double-HA tag and an EPEC null mutant complemented with the HA-tagged protein were differentially labeled, and lysates were mixed in a 1:1 ratio. Proteins were immunoprecipitated with the α-HA antibody and then trypsinized and analyzed by LC-MS/MS. (B) Representative mass spectra for peptides recovered from the SILAC experiment performed with EscL-2HA. Right panel: open and filled triangles indicate the expected mass charge (m/z) of light and heavy forms, respectively, of a peptide (QVIDQPFILGVR) from the specifically bound EscN from pulldowns with EscL-2HA and 2HA, respectively. The light peptide of EscN is present in a ratio >3.0 higher than the heavy peptide, indicating a specific interaction with EscL. Left panel: open and filled triangles indicate the expected m/z of light and heavy forms, respectively, of a peptide (TAESESSLMLR) from the nonspecifically bound EfeB protein from pulldowns with EscL-2HA and 2HA, respectively. (C) Summary of LEE-encoded proteins identified by LC-MS/MS as specifically immunoprecipitated with EscL-2HA, EscN-2HA, or EscQ-2HA in the SILAC experiments. H/L ratios represent the average values for the heavy/light isotope ratio that were determined among several peptides from either two or three independent immunoprecipitation experiments. S. D., standard deviations.