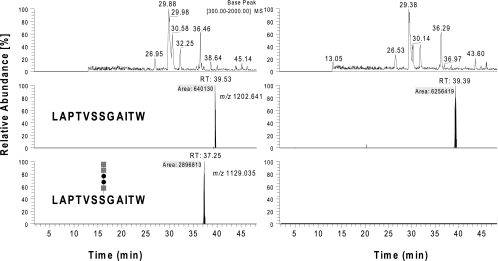
Fig. 8.
Characterization of PilA purified from type B strains FSC749 (wild type [left panels]) and FSC703 (ΔpglA background [right panels]). Base peak and selected ion chromatograms of chymotrypsin digests of PilA isolated from the two backgrounds were analyzed by nanoflow on-line LC-MS and manually inspected for glycan-related reporter ions and known precursor ions corresponding to modified PilA peptides. The top panel represents the base peak chromatograms (BPC), and the middle and bottom panels show the selected ion (SIC) chromatograms of precursor ions at m/z 1,202.6411+ (unmodified peptide 97LAPTVSSGAITW108) and it pentasaccharide modified form at m/z 1,129.0352+, respectively. Only the sample from the wild-type background showed a peak at 37.25 min related to the pentasaccharide modified peptide 97LAPTVSSGAITW108. Corresponding MS2 analysis confirmed the detection of the earlier described glycopeptides (see Fig. S2 in the supplemental material). The peak related to the unmodified peptide was detected at 39.4 to 39.5 min in both backgrounds (middle panels). Their abundance suggests that the concentration of the unmodified peptide in the sample from the ΔpglA background was approximately 10 times higher than that seen in the sample from the wild-type background (gray squares, HexNAc; filled circles, Hex).