Abstract
We previously found that enveloped virus binding and penetration are necessary to initiate an interferon-independent, IRF3-mediated antiviral response. To investigate whether membrane perturbations that accompany membrane fusion-dependent enveloped-virus entry are necessary and sufficient for antiviral-state induction, we utilized a reovirus fusion-associated small transmembrane (FAST) protein. Membrane disturbances during FAST protein-mediated fusion, in the absence of additional innate immune response triggers, are sufficient to elicit interferon-stimulated gene induction and establishment of an antiviral state. Using sensors of membrane disruption to activate an IRF3-dependent, interferon-independent antiviral state may provide cells with a rapid, broad-spectrum innate immune response to enveloped-virus infections.
TEXT
Mammalian hosts have evolved very complex mechanisms for recognizing and responding to incoming viral pathogens to limit their further replication and spread. These innate immune responses rely on a set of pathogen recognition receptors that recognize specific features of viruses and activate several signal transduction pathways, leading to the induction of antiviral responses. Following entry of viruses into cells, viral nucleic acids (DNA or RNA) are recognized by members of the Toll-like receptor (TLR)/RIG-I-like receptor (RLR)/nucleotide binding domain and leucine-rich repeat-containing (NLR) families, leading to the activation of the transcription factor interferon (IFN) regulatory factor 3 (IRF3) and production of type I IFNs (12, 18). These IFNs in turn engage alpha/beta IFN (IFN-α/β) receptors on neighboring cells, leading to a signal transduction cascade involving the Janus kinase and signal transducer and activator of transcription (Jak/STAT) pathway and the expression of numerous interferon-stimulated genes (ISGs) that function to limit virus spread (24). Although IRF3 is a major transcription factor responsible for IFN production, mounting evidence suggests that there are IRF3-independent mechanisms of IFN production in response to nucleic acid recognition (1, 8, 9, 20).
Recent evidence also suggests that cells can mount an IRF3-dependent, but IFN-independent, innate immune response. Enveloped, but not nonenveloped, virus particles from a broad range of virus families induce a subset of ISGs in the absence of detectable levels of viral replication and IFN production (3, 5, 13). Triggering this IFN-independent antiviral response requires binding to and penetration of the cell by the physical virus particle (13, 15, 16). Although IRF3 is essential for the response to virus particle entry (5, 17), upstream sensors of viral infection, including TLRs and RLRs, are not (19, 27). These data suggest that membrane fusion during enveloped-virus entry may be a sufficient trigger to induce an antiviral response. Indeed, cell-cell fusion of primary human fibroblasts mediated by expression of the fusogenic reptilian reovirus p14 fusion-associated small transmembrane (FAST) protein induces ISG56, MxB, and IP-10 expression (11). However, formal proof that membrane perturbation is sufficient to induce ISG expression has been lacking, due to the use of enveloped-virus particles or DNA plasmid expression systems in previous investigations. Given that viral and cellular RNA species are nonspecifically packaged into virus particles (14, 23, 25) along with the growing collection of cellular RNA and DNA sensors (28–30), the possibility of nucleic acid recognition following either virus particle entry or transfection with the p14 expression plasmid could not be excluded as a mechanism of IRF3 activation and ISG induction.
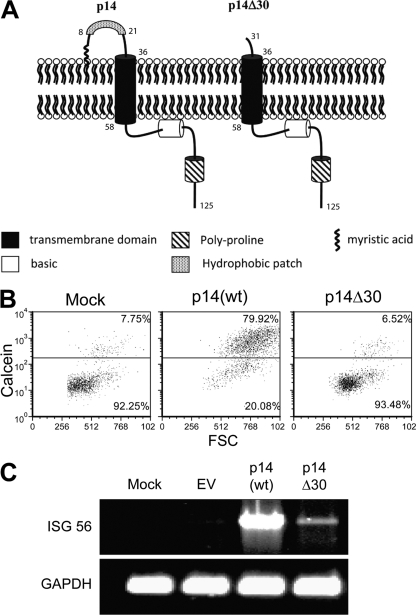
To directly address whether perturbations of the plasma membrane are sufficient to induce an antiviral state, we utilized wild-type (wt) and mutant forms of the p14 protein to induce cell-cell fusion or membrane perturbations (Fig. 1 A). The 125-residue p14 FAST protein is a nonstructural reovirus protein whose expression and localization to the plasma membrane induce cell fusion and syncytium formation (6, 10). To exclude the possibility that undefined innate triggers present in the p14 protein, rather than p14-induced membrane perturbations, trigger an ISG response, a mutant of p14 lacking part of an ∼36-residue ectodomain (p14Δ30) was included as a control. This mutant lacks the ectodomain hydrophobic patch that was previously shown to be required for syncytium formation (7). A cell-cell pore formation assay was utilized to quantify the percentage of donor cells coexpressing enhanced green fluorescent protein (eGFP) and p14 that acquired the cytoplasmic dye calcein red from the nontransfected target cells (4). While p14(wt) induced extensive cell-cell fusion, p14Δ30 was devoid of pore formation activity (Fig. 1B). Expression of p14(wt) in transiently transfected human fibroblasts induced the robust expression of ISG56, which is consistent with previous data (11), while expression of p14Δ30 had minimal effects on the induction of ISG56 (Fig. 1C).
Fig. 1.
p14(wt), but not the ectodomain mutant p14Δ30, is capable of inducing pore formation and ISG 56 mRNA accumulation. (A) Schematic diagram of the important domains within the p14 proteins. The p14Δ30 mutant lacks the first 30 NH2-terminal amino acids, including the hydrophobic patch. (B) The extent of pore formation was measured by adding calcein red-labeled Vero cells to QM5 cells cotransfected with eGFP and p14(wt) or p14Δ30 for 4 h. Pore formation was quantified by analyzing the percentage of gated eGFP-expressing cells that acquired the calcein red from the Vero cells, plotted against the forward scatter (FSC). (C) Vero cells transfected with pcDNA3.1 (EV), p14(wt), or p14Δ30 plasmid DNA were washed, trypsinized, and placed onto naïve human embryonic lung fibroblasts. RNA was harvested at 16 h post-cell transfer, followed by RT-PCR using primers specific for human ISG56 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
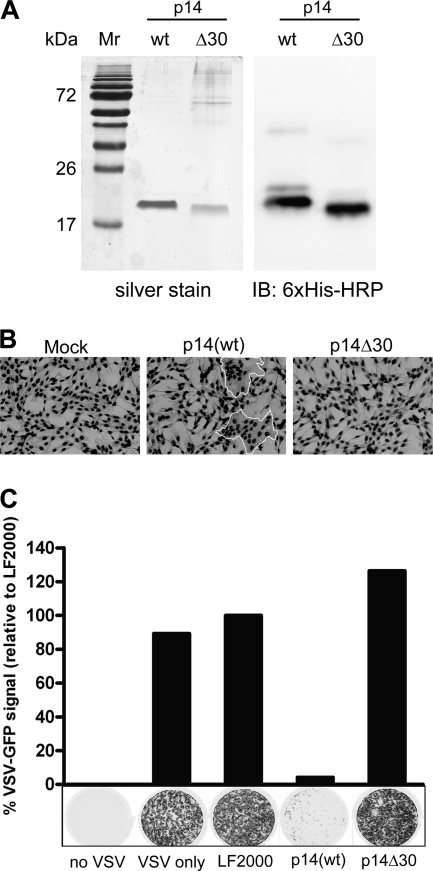
To eliminate any potential activation from plasmid DNA, the p14 proteins were expressed using baculovirus constructs in Sf21 insect cells and were purified by affinity and ion exchange chromatography as previously described (26) (Fig. 2 A). As these proteins are integral membrane proteins, they rapidly precipitate when diluted out of detergent and into culture medium. However, when diluted in the presence of a lipid carrier, p14 associates with the lipid vesicles and maintains its membrane fusion activity, as shown by the ability of purified p14(wt) in the presence of Lipofectamine 2000 (Invitrogen) to induce syncytium formation when added to fibroblasts (Fig. 2B). Consistent with the absence of pore formation activity (Fig. 1B), purified p14Δ30 was incapable of inducing syncytium formation when added to cells in the presence of Lipofectamine (7) (Fig. 2B). To determine whether the membrane fusion activity of purified p14 is sufficient to trigger a cellular antiviral response, primary human fibroblasts were treated with p14(wt) or p14Δ30 in the presence of Lipofectamine 2000. Treated cells were subsequently challenged with vesicular stomatitis virus (VSV), an RNA virus that is exquisitely sensitive to host innate responses, in a standard antiviral plaque reduction assay (13). Purified, fusion-active p14(wt) induced a robust antiviral state in these cells, a response that was not observed with treatment of cells with Lipofectamine alone (LF2000 control) or with Lipofectamine plus the nonfusogenic p14Δ30 protein (Fig. 2C). As was previously shown with enveloped virus particles (13), the induction of an antiviral state by purified p14(wt) occurred in the absence of detectable amounts of biologically active IFN (data not shown and Table 1). In addition, we failed to detect any signs of toxicity or cell death associated with these treatments (data not shown).
Fig. 2.
p14(wt)-Mediated cell-cell fusion induces an antiviral state in human fibroblasts. (A) Purification of p14 proteins following ion exchange chromatography. One microgram each of p14(wt) and p14Δ30 was run on a 12% SDS-polyacrylamide gel and silver stained. For Western immunoblot analysis, 100 ng each of p14(wt) and p14Δ30 was run on a 12% SDS-polyacrylamide gel and transferred onto a polyvinylidene difluoride (PVDF) membrane. A horseradish peroxidase-conjugated 6×His antibody was used to detect the p14 proteins. Mr, molecular mass marker. (B) QM5 cells were transfected with 4 μg of purified p14(wt) or p14Δ30 proteins for 8 h to allow fusion to proceed. Cells were fixed and Giemsa stained, and light microscopy images were captured at a magnification of ×200. Syncytia are outlined in white. (C) Following a 24-h treatment with purified p14(wt) or p14Δ30 proteins, human fibroblasts were infected with VSV expressing GFP from the viral promoter. GFP fluorescence was detected 24 h postinfection using a Typhoon Trio imager (GE Healthcare). Results from a representative experiment are presented. The level of GFP expression was quantified using ImageQuant TL software (GE Healthcare) and expressed as a percentage of fluorescence relative to that of LF2000-treated wells. Cells incubated with no VSV or VSV only were included as controls.
Table 1.
Activation of a subset of IFN-inducible genes following treatment of primary human fibroblasts with p14(wt) and p14Δ30a
| Functional gene group | GenBank accession no. | Gene | Description of gene product | Change in expression relative to that of LF2000 with indicated treatment |
||
|---|---|---|---|---|---|---|
| Mock | p14(wt) | p14Δ30 | ||||
| IFN-inducible genes | NM_001111 | ADAR | Adenosine deaminase, RNA specific | −1.02 | 2.09 | 1.37 |
| NM_001565 | CXCL10 | Chemokine (C-X-C motif) ligand 10 | −3.39 | 12.70 | 1.22 | |
| NM_005101 | ISG15 | ISG15 ubiquitin-like modifier | −1.29 | 21.85 | 2.31 | |
| NM_002038 | IFI6 | IFN-α-inducible protein 6 | −1.46 | 150.55 | 9.81 | |
| NM_005531 | IFI16 | IFN-γ-inducible protein 16 | −1.14 | 2.50 | 1.40 | |
| NM_005532 | IFI27 | IFN-α-inducible protein 27 | −1.17 | 114.04 | 6.86 | |
| NM_006332 | IFI30 | IFN-γ-inducible protein 30 | −1.12 | 2.15 | −1.08 | |
| NM_005533 | IFI35 | IFN-induced protein 35 | −1.04 | 5.76 | 1.41 | |
| NM_006417 | IFI44 | IFN-induced protein 44 | −1.04 | 10.52 | 2.28 | |
| NM_006820 | IFI44L | IFN-induced protein, 44-like | −1.59 | 151.35 | 9.14 | |
| NM_022168 | IFIH1 | IFN-induced protein with helicase C domain 1 | −1.78 | 10.28 | 2.21 | |
| NM_001548 | IFIT1 | IFN-induced protein with tetratricopeptide repeats 1 | 1.09 | 60.74 | 6.61 | |
| NM_001547 | IFIT2 | IFN-induced protein with tetratricopeptide repeats 2 | −1.03 | 8.62 | 2.18 | |
| NM_001549 | IFIT3 | IFN-induced protein with tetratricopeptide repeats 3 | −1.19 | 8.66 | 1.76 | |
| NM_003641 | IFITM1 | IFN-induced transmembrane protein 1 (9-27) | −1.03 | 20.31 | 2.64 | |
| NM_005373 | MPL | Myeloproliferative leukemia virus oncogene | 2.60 | 4.46 | 4.70 | |
| NM_002462 | MX1 | Myxovirus (influenza virus) resistance 1, IFN-inducible protein p78 | 1.87 | 538.21 | 25.21 | |
| NM_002534 | OAS1 | 2′,5′-Oligoadenylate synthetase 1, 40/46 kDa | 1.24 | 1178.69 | 41.36 | |
| NM_004509 | SP110 | SP110 nuclear body protein | −1.01 | 2.80 | 1.46 | |
| IFNs | NM_020124 | IFNK | IFN-κ | 1.24 | 4.53 | 1.81 |
| IFN receptors | NM_002187 | IL12B | Interleukin-12B (natural killer cell stimulatory factor 2) | 1.18 | 3.92 | 1.39 |
| NM_005373 | MPL | Myeloproliferative leukemia virus oncogene | 2.60 | 4.46 | 4.70 | |
| IFN regulatory factors | NM_002460 | IRF4 | IFN regulatory factor 4 | 1.24 | 2.46 | −1.26 |
| NM_001572 | IRF7 | IFN regulatory factor 7 | 1.26 | 6.77 | 1.54 | |
RT2 Profiler IFN pathway-specific arrays were utilized to investigate RNA expression profiles following treatment with p14. Human fibroblasts were treated with 4 μg of purified p14(wt) or p14Δ30 protein in the presence of LF2000 for 24 h. Genes whose expression was upregulated more than a factor of 2 relative to that of the LF2000 control in an experimental treatment group were identified.
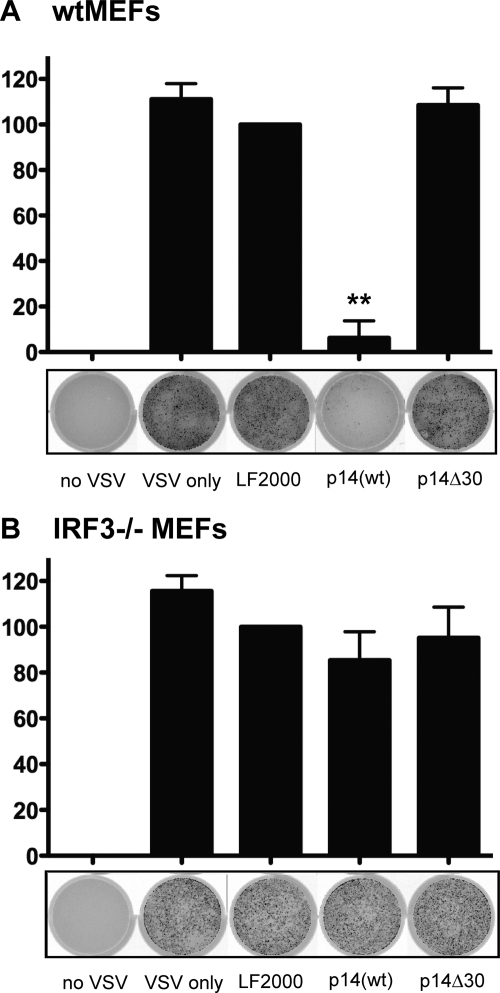
To investigate whether the antiviral state elicited by p14(wt) was dependent on IRF3, both wild-type and IRF3-deficient primary mouse fibroblasts were treated with the purified p14 proteins. Similar to p14(wt) treatment of primary human fibroblasts, treatment of wild-type mouse fibroblasts with the p14(wt) protein reduced the level of VSV-GFP expression by almost 90% relative to that of the LF2000 control (Fig. 3 B). Similar treatment of cells with the nonfusogenic p14Δ30 protein failed to induce an antiviral state in wild-type mouse fibroblasts (Fig. 3A and B). Most importantly, induction of the antiviral state by the p14(wt) protein was dependent on IRF3, since IRF3-deficient cells treated with p14(wt) protein were unable to prevent VSV-GFP replication. Collectively, these data demonstrate that membrane perturbation mediated by the p14 viral fusogen is sufficient to induce an IRF3-dependent, IFN-independent antiviral response in human and mouse cells.
Fig. 3.
p14(wt) triggers an antiviral state that is dependent on IRF3. Following a 24-h treatment with 4 μg of purified p14(wt) or p14Δ30 protein, wild-type (wt) (A) and IRF3−/− (B) MEFs were infected with VSV expressing GFP from the viral promoter. GFP fluorescence was detected 24 h postinfection using a Typhoon Trio imager (GE Healthcare). The level of GFP expression was quantified using ImageQuant TL software (GE Healthcare) and expressed as a percentage of fluorescence relative to that of LF2000-treated wells. Data are presented as means ± standard errors of the means (SEM) from three independent experiments. Statistical analysis was performed by one-way analysis of variance (ANOVA) and Tukey's post hoc test, comparing all treatments to that with LF2000 alone. Cells incubated with no VSV or with VSV only were included as controls. **, P > 0.001.
To investigate the RNA expression profile following treatment of primary fibroblasts with purified p14(wt) or p14Δ30 proteins, we utilized RT2 Profiler IFN pathway-specific arrays (Qiagen) consisting of 84 genes within four major functional groups (IFNs, IFN receptors, IFN regulatory factors, and ISGs). Genes whose expression was upregulated more than a factor of 2 relative to that of the LF2000 control in any experimental treatment group were identified (Table 1). Following treatment with the p14(wt) protein, 23 genes showed increased expression at the mRNA level. In marked contrast, there was a dramatic decrease in the induction levels of all 23 of these genes when cells were treated with the nonfusogenic p14Δ30 protein (Table 1). The complete data set from the PCR array may be found in Table S1 in the supplemental material. The gene induction profile induced by p14(wt) was similar to that obtained using a cDNA microarray following treatment of human fibroblasts with intact virus particles (13). Interestingly, of the 19 IFN genes on the array, only IFN-κ was moderately increased by p14(wt).
The results presented here strongly suggest that membrane perturbation is sufficient for induction of an innate cellular antiviral response. The cellular response elicited by membrane perturbations induced by the purified p14(wt) membrane fusion protein mirrors that observed upon virus particle entry: induction of a subset of ISGs in the absence of IFN production that is dependent on IRF3. The fact that the same response can be induced by a purified viral fusion protein indicates that this response is not triggered by virus pattern-associated molecular patterns such as nucleic acid. While we cannot definitively exclude the possibility that features of the p14 FAST protein beyond its membrane fusion activity might contribute to triggering this response, this seems highly unlikely since the p14Δ30 protein triggered no such response. Membrane localization studies (microscopy and biochemical fractionation) confirmed the cell surface localization of the utilized FAST proteins (data not shown). We are also unaware of known mechanisms of foreign (viral) proteins activating intracellular innate immune signaling events. Although viral glycoproteins engage cell surface receptors, including TLR2 and TLR4, p14 lacks high-affinity receptor binding activity (22). Furthermore, viral particle binding to the cell surface is insufficient to elicit the IFN-independent antiviral response (13, 15, 21). We are therefore left with the only reasonable conclusion, that the mechanism of p14-induced membrane fusion elicits induction of an antiviral response, the same response triggered by the entry of numerous enveloped, but not nonenveloped, viruses. Membrane perturbation associated with bacterial infection is also a known inducer of intracellular NLR proteins, leading to activation of the inflammasome (2), although the signaling pathways triggered in response to these perturbations have not been defined. With respect to viral entry, it remains to be determined what biochemical aspects of membrane fusion mediated by p14 or enveloped-virus fusogens lead to the types of membrane perturbations that trigger the observed cellular antiviral response. The signaling cascades upstream of IRF3 also remain elusive, as TLRs and RLRs were found to be nonessential (19, 27), consistent with the response being independent of nucleic acid or other viral structural components. It seems likely that membrane perturbations triggered by enveloped-virus entry are detected as a “cellular stress,” leading to rapid activation of a generic stress response pathway. Future experiments aimed at identifying the pathways involved should provide a clearer understanding of the primary events associated with a viral infection.
Supplementary Material
Acknowledgments
We thank B. Lichty for reagents, Jingyun Shou for expert technical assistance, and S. Dewitte-Orr for helpful discussions and critical reviews of the manuscript.
This work was supported by grants from the Canadian Institutes of Health Research to R.D. and K.L.M. R.S.N., K.T., and M.C. were supported by Natural Sciences and Engineering Research Council postgraduate scholarships.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 3 August 2011.
REFERENCES
- 1. Bourne N., et al. 2007. Early production of type I interferon during West Nile virus infection: role for lymphoid tissues in IRF3-independent interferon production. J. Virol. 81:9100–9108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brodsky I. E., Monack D. 2009. NLR-mediated control of inflammasome assembly in the host response against bacterial pathogens. Semin. Immunol. 21:199–207 [DOI] [PubMed] [Google Scholar]
- 3. Browne E. P., Wing B., Coleman D., Shenk T. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clancy E. K., Barry C., Ciechonska M., Duncan R. 2010. Different activities of the reovirus FAST proteins and influenza hemagglutinin in cell-cell fusion assays and in response to membrane curvature agents. Virology 397:119–129 [DOI] [PubMed] [Google Scholar]
- 5. Collins S. E., Noyce R. S., Mossman K. L. 2004. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J. Virol. 78:1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corcoran J. A., Duncan R. 2004. Reptilian reovirus utilizes a small type III protein with an external myristylated amino terminus to mediate cell-cell fusion. J. Virol. 78:4342–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Corcoran J. A., et al. 2004. Myristoylation, a protruding loop, and structural plasticity are essential features of a nonenveloped virus fusion peptide motif. J. Biol. Chem. 279:51386–51394 [DOI] [PubMed] [Google Scholar]
- 8. Daffis S., Suthar M. S., Szretter K. J., Gale M., Jr., Diamond M. S. 2009. Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 5:e1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DeWitte-Orr S. J., et al. 2009. Long double-stranded RNA induces an antiviral response independent of IFN regulatory factor 3, IFN-beta promoter stimulator 1, and IFN. J. Immunol. 183:6545–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duncan R., Corcoran J., Shou J., Stoltz D. 2004. Reptilian reovirus: a new fusogenic orthoreovirus species. Virology 319:131–140 [DOI] [PubMed] [Google Scholar]
- 11. Hancock M. H., Mossman K. L., Smiley J. R. 2009. Cell fusion-induced activation of interferon-stimulated genes is not required for restriction of a herpes simplex virus VP16/ICP0 mutant in heterokarya formed between permissive and restrictive cells. J. Virol. 83:8976–8979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawai T., Akira S. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143:1–20 [DOI] [PubMed] [Google Scholar]
- 13. Mossman K. L., et al. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muriaux D., Mirro J., Harvin D., Rein A. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U. S. A. 98:5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Netterwald J. R., et al. 2004. Postattachment events associated with viral entry are necessary for induction of interferon-stimulated genes by human cytomegalovirus. J. Virol. 78:6688–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nicholl M. J., Robinson L. H., Preston C. M. 2000. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J. Gen. Virol. 81:2215–2218 [DOI] [PubMed] [Google Scholar]
- 17. Noyce R. S., Collins S. E., Mossman K. L. 2009. Differential modification of interferon regulatory factor 3 following virus particle entry. J. Virol. 83:4013–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Neill L. A., Bowie A. G. 2010. Sensing and signaling in antiviral innate immunity. Curr. Biol. 20:R328–R333 [DOI] [PubMed] [Google Scholar]
- 19. Paladino P., Cummings D. T., Noyce R. S., Mossman K. L. 2006. The IFN-independent response to virus particle entry provides a first line of antiviral defense that is independent of TLRs and retinoic acid-inducible gene I. J. Immunol. 177:8008–8016 [DOI] [PubMed] [Google Scholar]
- 20. Prescott J. B., Hall P. R., Bondu-Hawkins V. S., Ye C., Hjelle B. 2007. Early innate immune responses to Sin Nombre hantavirus occur independently of IFN regulatory factor 3, characterized pattern recognition receptors, and viral entry. J. Immunol. 179:1796–1802 [DOI] [PubMed] [Google Scholar]
- 21. Preston C. M., Harman A. N., Nicholl M. J. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909–8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salsman J., Top D., Barry C., Duncan R. 2008. A virus-encoded cell-cell fusion machine dependent on surrogate adhesins. PLoS Pathog. 4:e1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sciortino M. T., Suzuki M., Taddeo B., Roizman B. 2001. RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol. 75:8105–8116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sen G. C., Sarkar S. N. 2007. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr. Top. Microbiol. Immunol. 316:233–250 [DOI] [PubMed] [Google Scholar]
- 25. Terhune S. S., Schroer J., Shenk T. 2004. RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration. J. Virol. 78:10390–10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Top D., et al. 2005. Liposome reconstitution of a minimal protein-mediated membrane fusion machine. EMBO J. 24:2980–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsitoura E., et al. 2009. Infection with herpes simplex type 1-based amplicon vectors results in an IRF3/7-dependent, TLR-independent activation of the innate antiviral response in primary human fibroblasts. J. Gen. Virol. 90:2209–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Unterholzner L., et al. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11:997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang P., et al. 2010. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat. Immunol. 11:487–494 [DOI] [PubMed] [Google Scholar]
- 30. Yoneyama M., Fujita T. 2010. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 20:4–22 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.