Abstract
The encephalomyocarditis virus (EMCV), a Picornaviridae virus, has a wide host spectrum and can cause various diseases. EMCV virulence factors, however, are as yet ill defined. Here, we demonstrate that the EMCV 2A protein is essential for the pathogenesis of EMCV. Infection of mice with the B279/95 strain of EMCV resulted in acute fatal disease, while the clone C9, derived by serial in vitro passage of the B279/95 strain, was avirulent. C9 harbored a large deletion in the gene encoding the 2A protein. This deletion was incorporated into the cDNA of a pathogenic EMCV1.26 strain. The new virus, EMCV1.26Δ2A, was capable of replicating in vitro, albeit more slowly than EMCV1.26. Only mice inoculated with EMCV1.26 triggered death within a few days. Mice infected with EMCV1.26Δ2A did not exhibit clinical signs, and histopathological analyses showed no damage in the central nervous system, unlike EMCV1.26-infected mice. In vitro, EMCV1.26Δ2A presented a defect in viral particle release correlating with prolonged cell viability. Unlike EMCV1.26, which induced cytopathic cell death, EMCV1.26Δ2A induced apoptosis via caspase 3 activation. This strongly suggests that the 2A protein is required for inhibition of apoptosis during EMCV infection. All together, our data indicate that the EMCV 2A protein is important for the virus in counteracting host defenses, since Δ2A viruses were no longer pathogenic and were unable to inhibit apoptosis in vitro.
INTRODUCTION
The encephalomyocarditis virus (EMCV) belongs to the genus Cardiovirus, within the family Picornaviridae, which also contains the better known genera Enterovirus and Aphthovirus. EMCV is a nonenveloped virus, with an icosahedric capsid and a genome consisting of a positive single-stranded RNA of about 7.8 kb linked to a viral protein (VPg) at its 5′ extremity. The EMCV RNA genome is composed of a unique coding sequence flanked by untranslated regions (UTR). The 5′ UTR comprises an internal ribosome entry site (IRES), which allows direct initiation of translation of the single viral polyprotein and, like aphthoviruses, a poly(C) tract. The 3′ UTR terminates with a heterogeneous poly(A) tail that interacts with the 3D protein, a viral RNA-dependent RNA polymerase (12).
EMCV proteins and precursors get their names from their position in the polyprotein, that is, the leader protein (L), the precursor P1, comprising capsid proteins VP4, VP2, VP3, and VP1, P2 and P3, precursors of the nonstructural proteins 2A, 2B, and 2C, and 3A, 3B (also called VPg), the 3C protease, and the RNA-dependent RNA polymerase 3D (39). Polyprotein cleavage is carried out mainly by the 3C protease of EMCV. However, the first “cleavage” occurs during elongation, prior to the synthesis of the 3C protease, between the 2A and 2B proteins, due to an inherent instability of the peptide chain at the NPG(P) sequence (22).
Unlike other picornaviruses, the 2A proteins of cardio- and aphthoviruses are not proteases. The EMCV 2A protein, containing 143 amino acids (aa), has been implicated in the virus-induced shutoff host protein synthesis often observed during picornavirus infection (6), since partial deletions in the 2A protein result in maintenance of cellular mRNA translation (47). During EMCV infection, the shutoff host protein synthesis is not as rapid or extensive as during poliovirus infection (24). Indeed, in the case of EMCV, it is thought to stem from competition between cap- and IRES-dependent translation (21) rather than be a definitive shutoff.
Several nonexclusive mechanisms by which the EMCV 2A protein may inhibit host protein synthesis have been proposed. Inhibition of cap-dependent translation could be due to the binding of 2A to eIF4E, which might impede the interaction of eIF4E with eIF4G (eIF4E-eIF4G-eIF4A interactions are required for initiation of cap-dependent translation). An eIF4E binding site between amino acids 126 to 134 has been located (20). Another possibility is the activation of an inhibitor of translation: when 2A was expressed in BHK-21 cells, 4E-BP1 was observed to be hypophosphorylated (47). Since 4E-BP1 is active when dephosphorylated and sequesters eIF4E, cap-dependent translation would be expected to be inhibited. This phenomenon, however, was not found in HeLa and L cell types (20). Early after infection, 2A is localized in the nucleoli, owing to its nuclear localization signal (NLS) (20), and associates with the nascent ribosomal subunit (2). Depletion of the NLS has been shown to diminish inhibition of cap-dependent translation. In the cytosol, the 2A protein interacts with the 40S ribosomal subunit (21, 33). This association is believed to induce preferential use of IRES-dependent templates (rather than capped mRNA).
EMCV is known as a cytolytic virus. When lysis (necrosis) occurs, the plasma membrane breaks down, allowing release of virions, cellular components, and proinflammatory molecules that can damage tissue and give rise to an inflammatory response (15). Apoptosis or programmed cell death is a common cell defense mechanism to diverse viral infections and represents, at a cellular scale, self-sacrifice. It is characterized by chromatin condensation, nuclear fragmentation, and formation of apoptotic bodies (15). The cascade of events responsible for these morphological changes is induced mainly by members of the caspase family. These caspases are present constitutively in cells but require proteolytic processing to become active. The cleaved form of caspase 3 is known to be an effector of apoptosis. Apoptotic cell death is thought to prevent generation and spread of viral progeny. Many viruses, however, have evolved to circumvent this defense mechanism. Intracellular inhibitors of apoptosis have been identified in polioviruses (4, 35), coxsackieviruses (16, 43), and cardioviruses (41). In regards the latter genus, induction of apoptosis has been described in certain cell types (44, 52, 55). Recently, however, the leader protein of Mengovirus (closely related to EMCV) has been found to mediate antiapoptotic activity (41). In keeping with the idea that EMCV mediates antiapoptotic activity, inhibition of programmed cell death has been shown to be required for EMCV virulence in mice (44).
EMCV displays a wide spectrum of host and disease, as it is able to infect nonhuman primates, swine, boars, rodents, and elephants, and human infections have also been reported (36). In mice, EMCV causes mainly myocarditis (11), neurovirulence (48–51), and diabetes (56).
The molecular determinants of EMCV virulence and pathogenicity are not fully understood. In mice, these have been investigated mainly for diabetes (5, 56). Here we show that deletion of 115 amino acids from the EMCV 2A protein of two different strains profoundly affects their virulence. Despite the deletion, the virus remained viable in vitro, albeit displaying a decrease in virus yield and a defect in particle release. Our data also indicate that cells infected with EMCV1.26Δ2A underwent apoptosis characterized by caspase 3 activation, unlike those infected with the wild-type virus, which did not exhibit apoptotic signs. These results strongly suggest that the 2A protein of EMCV is required for apoptosis inhibition, which may be important for viral pathogenesis.
MATERIALS AND METHODS
Cell culture and viruses.
The baby hamster kidney 21 (clone 13) (BHK-21) cell line was maintained in complete growth medium, as follows: minimal essential medium (MEM), 10% fetal bovine serum (FBS), 1% nonessential amino acids (NEAA), and 1% sodium pyruvate. Antibiotics, 1 U/ml of penicillin and 1 μg/ml of streptomycin, were added to all media. Cells were maintained in a 5% CO2 atmosphere at 37°C.
The highly virulent B279/95 and the attenuated B279/95p210 EMCV strains were kindly provided by Frank Koenen (Department of Virology, Section of Epizootic Diseases, CODA-CERVA, Groeselenberg 99, B-1180 Ukkel, Belgium) (13). Briefly, B279/95 was originally isolated from a piglet that died from myocarditis in a pig-fattening farm in Belgium in 1995. It has been passed 210 times in BHK-21 cells so as to obtain a strain attenuated for pigs, the B279/95p210 strain. The clone B279/95p210-C9 was isolated from B279/95p210 by plaque assay.
The EMCV1.26 and EMCV1.26Δ2 viruses were obtained following transfection of cells with RNAs transcribed from p1.26 and p1.26Δ2A plasmids, respectively (see below).
Plasmids.
Plasmid p1.26 (also called pBL/T7EMCgB2887) has been previously described (28). It contains the full-length cDNA of the Belgium EMCV 2887A/91 strain. This strain was originally isolated from a fetus obtained from a pig-breeding farm with a history of reproductive failure (14). Plasmid p1.26Δ2A was obtained by introduction of a deletion of 345 nucleotides (nt) (from nt 30 to 375) in the 2A protein coding sequence (see Fig. 2a). The deleted plasmid was engineered by a technique called seamless fusion of DNA fragment by overlap PCR (31) and subcloning. In the first step, two separate PCR were performed to amplify 2 fragments flanking the 5′ and 3′ extremities of the deletion. The negative-sense primer for the 5′ side fragment and the positive-sense primer for the 3′ side fragment are completely complementary to each other. The 5′ side fragment was 3,282 bp, and the 3′ side fragment 935 bp. These two PCR products were then used as templates for a second PCR amplification with a third couple of primers. The third positive-sense primer was located at the 5′ end of the large fragment, and the third negative-sense primer was located at the 3′ end of the small one. The final “fusion 5′-3′” product was digested with HindIII and SacII and cloned into a pBluescript SK(+) vector (Stratagene). To obtain the full-length genomic cDNA of the 2A-deleted virus, the HindIII-SacII fragment of the p1.26 plasmid was replaced by the corresponding fragment bearing a deletion in the 2A-encoding sequence.
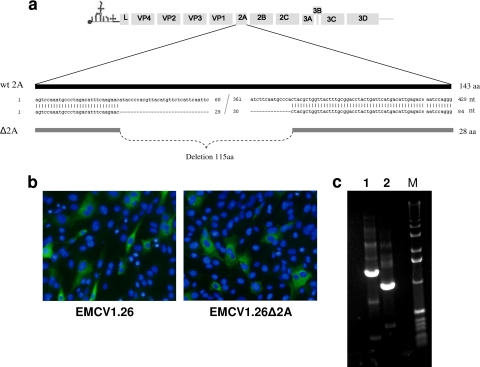
Fig. 2.
Production of EMCV1.26Δ2A. (a) Schema of the 2A deletion introduced into the EMCV cDNA p1.26. (b) BHK-21 cells were infected with EMCV1.26 or EMCV1.26Δ2A at an MOI of 1 for 5 h and immunostained with anti-EMCV VP1 (clone 3E5) followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (green) and DAPI (blue). (c) Electrophoresis of products amplified by RT-PCR of viral RNA of EMCV1.26 (1) and EMCV1.26Δ2A (2) using primers flanking the 2A sequence. M, TrackIt 1-kb DNA ladder; nt, nucleotides; aa, amino acids; wt, wild-type.
In vitro transcription and transfection.
Plasmids p1.26 and p1.26Δ2A were linearized by digestion with NotI and purified with the QIAquick purification kit (Qiagen). Genomic RNAs were transcribed from the linearized plasmids, using the MEGAscript in vitro transcription kits for large-scale synthesis of RNAs (Ambion) for 4 h at 37°C, and then treated with Turbo DNase for 15 min at 37°C, as recommended by the manufacturer. RNA integrity was examined by electrophoresis on agarose gels.
BHK-21 cells were transfected with RNAs transcribed from p1.26 or p1.26Δ2A by using TransFast transfection reagent (Promega), as recommended by the manufacturer. Briefly, growth medium was removed from subconfluent (90 to 95%) cells in 24-well culture plates. RNA (1 μg) was mixed with 3 μl of reagent in a final volume of 200 μl of MEM (Gibco) and applied to the cells for 1 h at 37°C. Complete growth medium was then added, and cells were incubated at 37°C. When more than 80% of the cells showed a cytopathic effect, they were subjected to one freezing and thawing cycle. The viral suspension was clarified by centrifugation for 10 min at 2,000 × g and stored at −80°C. Virus stocks of EMCV1.26 and EMCV1.26Δ2A were produced by 3 passages on BHK-21 cells.
In vitro infections and titration.
One-day cultures of subconfluent (90 to 95%) cultures of BHK-21 cells were used for infection. Two wells served to determine the number of cells/well and calculate the concentration of virus to add. The growth medium was discarded from cells to be infected, and the virus suspension was added after dilution (in a final volume of 50, 100, or 500 μl for 96-, 24-, or 6-well plates, respectively) to provide the desired multiplicity of infection (MOI). After 1 h at 4°C, cells were washed three times with cold MEM, and warm complete media were added. Cells were then incubated for the indicated time at 37°C. For the one-step growth curve, cells and supernatant were frozen together at −80°C at the indicated time points, while for comparison of free and cell-associated virus titers, supernatant and cells were harvested and frozen separately, and in this case, cell lysates were resuspended in MEM before titration.
Virus was quantified by endpoint dilution using 8 wells per virus dilution as previously described (46), with titers expressed as the median tissue culture infective doses per ml (TCID50/ml), or by plaque assays on BHK-21 cells. Cytopathic effects were evaluated 3 days after infection.
Measurement of cell viability.
The cell proliferation reagent WST-1 (Roche) was used to measure BHK-21 viability after infection. Cells were mock infected or infected with EMCV1.26 or EMCV1.26Δ2A at the indicated MOI in triplicate in 96-well plates. After 10 and 24 h of incubation, 20 μl/well of WST-1 was added. The absorbance at 450 nm was read just after adding the reagent and after incubation for 2 h at 37°C. The percentage of viability was calculated with reference to the optical density (OD) value obtained for uninfected cells, which assigned 100% viability.
Mice and inoculation.
All animal protocols were approved by the institutional guidelines for animal care. Mice were housed in an environmentally controlled room under biosafety level 3 conditions.
Female C57BL/6 mice, 4 to 6 weeks old, were purchased from Charles River Laboratories (Lyon, France) and infected intraperitoneally (i.p.) with 400 μl of diluted virus or MEM in 4 different experiments.
For the first experiment, groups of 20 mice were inoculated with 2.4 × 108 PFU of B279/95, B279/95p210, or B279/95p210-C9 virus. As a negative control, 5 mice were inoculated with MEM. Mice were monitored for 22 days with observation of clinical signs, including hunched posture, abnormal gait, ruffled fur, encephalitis, hind-limb paralysis, quadriplegia, and death.
In a second experiment involving the genetically engineered viruses EMCV1.26 and EMCV1.26Δ2A, described above, groups of five mice were inoculated with 4 × 105 PFU/mouse of each virus or, as a control, MEM, and evaluated clinically for 18 days.
In a third experiment, two groups of 35 mice were inoculated, one group with EMCV1.26 and the other group with EMCV1.26Δ2A. Following inoculation on day 0 with 4 × 104 PFU/mouse, five mice per day (days 1, 2, 3, 4, 7, 9, and 22) and per virus were randomly sacrificed. Tissues from mice that succumbed to EMCV infection were harvested on the day of death. Two untreated and three MEM-inoculated mice were euthanized at day 0 or at the end of the experiment, respectively, and served as negative controls. From each mouse, the heart, brain, and lumbar and thoracic spinal cords were recovered for histopathology or real-time reverse transcription-PCR (RT-PCR) analyses.
For the last experiment, 21 mice were i.p. inoculated with 8 × 106 TCID50 of EMCV1.26Δ2A, and 4 mice were inoculated with MEM. Three mice per indicated day (as in the third experiment) were randomly sacrificed. Their hearts and brains were harvested for real-time RT-PCR analyses.
Histopathological and immunohistochemical analyses.
Three brains and spinal cords were recovered from mice per virus and per day. Tissues were fixed in 10% neutral buffered formalin, dehydrated, and embedded in paraffin. Spinal cords were first embedded in agar gel to allow multiple transversal sections.
Six sections of the brain and two from both the thoracic and lumbar spinal cords were stained with hematoxylin and eosin and saffron and subjected to histopathological examination.
All immunohistochemical steps were performed on the Discovery apparatus (Roche), according to a previously described method with a primary antibody 3E5 at a dilution of 1/500 (25).
Real-time RT-PCR.
Tissue samples were crushed using Lysing Matrix D tubes with the FastPrep 24 equipment, and total RNA was extracted using the RNeasy minikit (Qiagen), according to the manufacturer's recommendation. Then, real-time RT-PCRs were performed. For EMCV quantification, an in-house standard and the QuantiTect probe RT-PCR kit (Qiagen) were used. For normalization with the 18S RNA, the QuantiTect SYBR green RT-PCR kit (Qiagen) and QuantiTect primer assay (Hs_RR18S_1_SG) (Qiagen) were used for each sample.
Primers and the TaqMan probe used for EMCV detection were located in the conserved sequence of EMCV 3D. Primers used were EMC-TMF1, 5′-GGGATCAGCTTTTACGGCTTT-3′, EMC-TMR1, 5′-TGCATCCGATAGAGAACTTAATGTCT-3′, and the TaqMan probe used, synthesized by Applied Biosystems, was EMCVTMFR1, 5′FAM-CGATGCCAACGAGGACGCCC- MGB 3′.
Real-time RT-PCR was carried out in a 20-μl reaction mixture. For EMCV quantification, 5 μl of RNA extract was mixed with 1× QuantiTect Probe RT-PCR master mix, 0.5 μM each EMCV-TMF1 and EMCV-TMR1, 1× QuantiTect RT mix, and 0.2 μM EMCVTMFR1 probe.
For normalization with 18S RNA, 5 μl of 1/1,000 diluted RNA extract was mixed with 1× QuantiTect SYBR green RT-PCR master mix, 1× primer Hs_RR18S_1_SG, and 1× QuantiTect RT mix supplemented with RNase-free water.
A LightCycler (Roche) system was used to perform real-time RT-PCR. Samples were held at 50°C for 20 min for RT and at 95°C for 15 min for Taq activation and then amplified by 45 cycles consisting of denaturation for 15 s at 94°C, hybridization for 20 s at 55°C, and elongation for 10 s at 72°C. The protocol was ended by a cooling step for 30 s at 40°C. Results were analyzed using the LightCycler software.
For quantification, standard curves were generated by amplification of serial dilutions of known concentrations of purified in vitro transcripts of EMCV RNA ranging from 5 × 106 to 2.5 × 101 copies. Data were normalized by expressing EMCV RNA as the ratio of EMCV RNA copies to specific cellular RNA.
Apoptosis induction.
Staurosporine (STS) (Sigma) was used as a nonviral inducer of apoptosis. Cells were incubated for 16 to 18 h at 37°C under a 5% CO2 atmosphere with 0.5 μM STS in complete medium.
Antibodies.
To detect EMCV, an anti-EMCV monoclonal antibody (3E5) raised against the EMCV Novara 86 strain, obtained from mouse ascitic fluid, was used (kindly provided by Emiliana Brocchi, Instituto Zooprofilattico Sperimentale della Lumbaria e dell'Emilia B. Ubertini, Brescia, Italy). It recognizes a linear epitope on VP1 and cross-reacts with VP2 and VP3 (54). To detect apoptotic cells, a rabbit polyclonal antibody directed against the cleaved caspase 3 (Asp175) antibody (Cell Signaling) was used.
The secondary antibodies used were as follows: goat anti-mouse IgG coupled with phycoerythrin (PE) (5889-500; Abcam); goat anti-mouse IgG coupled with Alexa 546 (Invitrogen/Molecular Probes); Alexa Fluor 488-conjugated goat anti-rabbit IgG (A-11008; Invitrogen/Molecular Probes), and for flow cytometry, Alexa 647-conjugated anti-mouse IgG (A21236; Invitrogen/Molecular Probes).
Immunofluorescence.
Immunofluorescence (IF) assays were used to detect viral protein expression and cleaved caspase 3. BHK-21 cells were infected with wild-type and deleted 2A virus at an MOI of 10, as indicated above, and incubated for the indicated time. After discarding the supernatant, cells were fixed for 10 min at −20°C with cold 90% acetone or using 4% paraformaldehyde (PFA) for 20 min at room temperature (RT°). The fixed monolayer cells were washed twice with phosphate-buffered saline (PBS) and incubated for 30 min at 37°C with PBS-0.3%Triton-3% bovine serum albumin (BSA). Cells were then incubated with the 3E5 anti-EMCV monoclonal antibody and the rabbit anti-cleaved caspase 3 polyclonal antibody diluted to 1:200 in PBS-0.1%Triton-0.3% BSA for 1.5 h at RT°. After three washes with PBS-0.1% Triton-0.3% BSA, cells were incubated for 1 h at RT° in the dark with PE-conjugated goat anti-mouse IgG and Alexa Fluor 488-conjugated goat anti-rabbit IgG, both at a 1:1,000 dilution. After 3 washes with PBS-0.1% Triton-0.3% BSA, cells were washed once more with PBS, and DNA was stained for 20 min at RT° with 4′,6-diamidine-2-phenylindole dihydrochloride (DAPI) (Roche), followed by 3 washes with PBS. For cells plated on Lab-Tek chamber slides (134941; Lab-Tek Brand Products), a final wash with water was performed before mounting slides using Shandon Immu-Mount (9990412; Thermo Scientific). Pictures were taken using an Apotome microscope (10211786540; Zeiss) and AxioVision software.
Labeling of DNA fragmentation characteristic of apoptosis was performed, using the DeadEnd fluorometric terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) system (G3250; Promega), on mock cells, STS-treated cells, and EMCV1.26- and EMCV1.26Δ2A-infected cells (MOI of 10). The TUNEL staining was performed as recommended by the manufacturer.
Detection of apoptosis and EMCV infection by flow cytometry.
BHK-21 cells were seeded in 6-well plates a day prior to infection at an MOI of 5 (procedure described in detail above) or incubated for 18 h with STS for induction of apoptosis. At 8 and 14 h after infection, cells were resuspended and centrifuged at 300 × g (as for all following centrifugation steps). Supernatants were discarded, and cells fixed with PFA 4% for 20 min at RT°. Permeabilization and blocking were performed with PBS-0.3% Triton-3% BSA for 45 min at RT°, followed by incubation with anti-cleaved caspase 3 antibody (dilution 1/300), anti-EMCV (3E5) (dilution 1/200) in PBS-0.1% Triton-0.3% BSA for 1.5 h at RT°. Secondary antibodies were incubated for 1 h using the same dilution buffer. Cells were washed three times before and after incubation with secondary antibodies. A final wash was performed with PBS just before resuspension in PBS. Cells were analyzed immediately by flow cytometry (FACS Canto II; BD Biosciences) for cleaved caspase 3-Alexa Fluor 488 and VP1 EMCV-Alexa 647 binding. For each sample, data from 10,000 cells were collected. Dot plots and histograms were analyzed by FlowJo V.7.6.1 software (Tree Star, Inc.). Cleaved caspase 3-positive cells were considered apoptotic, and EMCV VP1-positive cells were considered infected.
Translation inhibition.
Rapamycin (Sigma) was used as an inhibitor of cap-dependent translation (7, 47). Cells were incubated with 100 nM rapamycin in complete medium for 24 h and then transfected with the plasmid pRSV-βGal, as previously described (8), as a reporter of cap-dependent translation. Plasmid transfections were performed using JetPrime DNA transfection reagent (Polyplus Transfection), briefly, for a well of a 24-well plate: 500 ng of pRSV-βGal was diluted with 50 μl of JetPrime buffer and 1 μl of JetPrime reagent, set for 10 min, and added to cell media containing or not containing rapamycin. A total of 44 h later, supernatants were discarded, and cells were lysed using a specific buffer that allows following measurement of the β-galactosidase (β-Gal) activity as described by Breiman et al. (8). The β-galactosidase activity is directly proportional to its production and thus to the cap-dependent translation activity. Absorbance was read at 450 nm.
Rapamycin cells induced for 24 h were also used for infection with EMCV1.26Δ2A at an MOI of 10. At the indicated time, those cells were fixed and immunostained for cleaved caspase 3 and EMCV-VP1 as described in “Immunofluorescence.”
RESULTS
I. Identification of an attenuated EMCV.
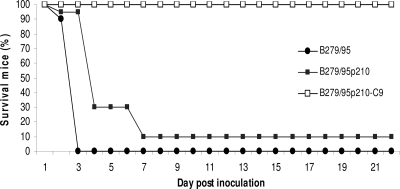
In order to identify molecular determinants of EMCV virulence in mice, we began our investigation by isolating, using plaque assay, clones from an attenuated strain of EMCV, B279/95p210. The attenuated virus was obtained by 210 passages of the highly virulent Belgian isolate B279/95 on BHK-21 cells and was demonstrated to no longer be pathogenic for pigs (13).
To evaluate the virulence of B279/95, B279/95p210, and isolated clones, C57BL/6 mice were infected intraperitoneally (i.p.) with a high dose of virus and monitored for clinical signs for 22 days (Fig. 1). Signs of paralysis were observed as early as 2 days after inoculation for mice infected with either B279/95 or B279/95p210. Three days after inoculation, all animals infected with B279/95 had died, while 6 days were required for the group infected with B279/95p210 to reach 90% mortality. The fact that B279/95p210-infected mice showed delayed clinical signs and that 2 mice survived suggested that this virus was partially attenuated in mice.
Fig. 1.
Identification of an attenuated EMCV virus. C57BL/6 female mice (n = 60) were i.p. injected with 2.4 × 108 PFU of the virulent B279/95 (•) or attenuated B279/95p210 (▪) strain or the clone B279/95p210-C9 (□) and followed for 22 days.
Of note, animals infected with one of the clones isolated from B279/95p210 (clone B279/95p210-C9) did not present any clinical signs (Fig. 1).
Sequence comparison between B279/95 and the C9 clone revealed 2 mutations in the L protein, 8 mutations in capsid proteins, and a deletion of 115 amino acids in the 2A protein of this virus. This deletion was found only in the C9 clone, while the other mutations were found in other pathogenic clones (data not shown). In order to determine whether this deletion in the 2A protein of EMCV could, on its own, be responsible for the attenuation of virulence in mice, it was introduced into another virulent EMCV strain.
II. Production of EMCV1.26Δ2A.
The deletion of 345 nucleotides found in the B279/95p210-C9 clone was introduced into the 2A coding region of the infectious EMCV cDNA (clone p1.26) previously produced in our laboratory (27). The deletion comprised nucleotides 30 to 375 and is represented in Fig. 2 a. Genomic RNAs were transcribed in vitro from the cDNA of the EMCV deleted in the 2A region (p1.26Δ2A) and from the cDNA of the wild-type virus (p1.26) and used to transfect BHK-21 cells for production of recombinant viruses.
Transfection with both types of RNA gave rise to a cytopathic effect within 24 to 36 h. The infectivity of newly generated viruses was assessed by immunofluorescence staining for EMCV-VP1 at 5 h after inoculation of cell culture (Fig. 2b). At this time point, both viruses expressed VP1, suggesting that transfection with the EMCV1.26Δ2A RNA had generated infectious viruses. RT-PCR was performed on RNA extracted from the EMCV1.26Δ2A virus after three passages in cell culture to verify the presence of the 2A deletion. The RT-PCR amplified a sequence encompassing the 2A deletion. The bands obtained for EMCV1.26 and EMCV1.26Δ2A presented a difference of approximately 345 bp, corresponding to the 2A deletion introduced into EMCV1.26Δ2A (Fig. 2c).
These results also suggested that the integrity of the 2A protein was not required for viral infectivity in vitro.
III. Characterization of EMCV1.26Δ2A.
To characterize the EMCV1.26Δ2A virus, plaque sizes and growth kinetics of the Δ2A virus and its wild-type parent were compared (Fig. 3). Wild-type EMCV1.26 yielded large (approximately 8-mm) clear plaques, while the 2A mutant exhibited a small-plaque phenotype (1 to 2 mm) when assayed on BHK-21 monolayers (Fig. 3a).
Fig. 3.
Characterization of the attenuated EMCV1.26Δ2A virus. (a) Plaque assays monitored the phenotype of EMCV1.26 and EMCV1.26Δ2A in BHK-21 cells, 72 hpi. (b) One-step growth curves of EMCV1.26 (•) and EMCV1.26Δ2A (▴) on BHK-21 cells. An MOI of 10 was used for infection, and at each indicated time, supernatant and cells were harvested and freeze-thawed together for viral titration, as described in Materials and Methods.
The one-step growth curve of EMCV1.26Δ2A exhibited a 1-h extension of the latent phase and also reached a plateau 1 h later than the wild-type EMCV1.26 (Fig. 3b). At the peak of the curve, production of the Δ2A virus was approximately 1 log lower. Thus, EMCVΔ2A exhibited less efficient virus production and an attenuated phenotype in vitro.
IV. EMCV 2A protein is required for pathogenesis in vivo.
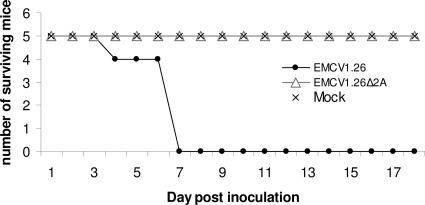
To evaluate the virulence of EMCV1.26 and EMCV1.26Δ2A in vivo, C57BL/6 female mice were infected intraperitoneally with 4 × 105 PFU of viruses. Signs of paralysis and death were observed as early as 4 days after inoculation in EMCV1.26-infected animals (Fig. 4). Seven days after infection, all animals had died. In contrast, no animal infected with EMCV1.26Δ2A died, and none exhibited any clinical signs.
Fig. 4.
The EMCV 2A protein is required for pathogenesis in vivo. Number of surviving C57BL/6 female mice (n = 15) i.p. injected with EMCV1.26 (•), EMCV1.26Δ2A (Δ), or MEM (×) during the 18-day follow-up are represented. Mice inoculated with EMCV1.26Δ2A did not exhibit clinical signs.
EMCV1.26Δ2A, like B279/95p210-C9, was nonpathogenic for mice (Fig. 1 and 4). Since both bear a deletion in the 2A protein, it appeared that the 2A protein is required for EMCV pathogenesis in vivo.
No external clinical signs were observed in EMCV1.26Δ2A-infected mice. To determine whether this virus can cause tissue damage, especially in the central nervous system (CNS), we repeated this experiment and harvested brains and thoracic and lumbar spinal cords on different days after infection. Histopathological examination of these tissues was performed, and representative pictures are shown in Fig. 5.
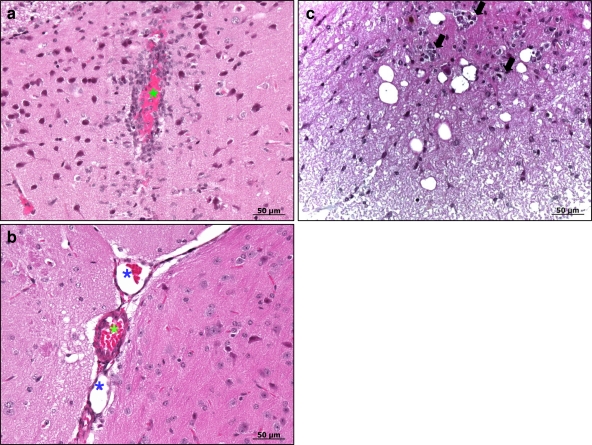
Fig. 5.
Histopathology of brains and spinal cords of C57BL/6 female mice i.p. injected with EMCV1.26 or EMCV1.26Δ2A. (a) Mononuclear perivascular infiltrate in the brain of a mouse infected by EMCV1.26 at 4 days postinfection. (b) Absence of any lesions in the brain of a mouse infected by EMCV1.26Δ2A. (c) In the spinal cords of EMCV1.26-inoculated mice, mononuclear infiltrates (arrows) have a smaller size than those in the brain. Venule (blue star); arteriole (green star). Hematoxylin counterstaining is shown.
One of the mice inoculated with EMCV1.26 exhibited discrete lesions in the brain but not in the spinal cord at day 3. At day 4, 3 mice exhibited lesions in brain and spinal cord, while at day 7 after infection, all of the mice inoculated with virus presented multifocal nervous lesions. These nervous lesions consisted of perivascular mononuclear infiltrate in the form of perivascular cuffs, multifocal neuronal necrosis, and diffuse gliosis. Cerebral lesions were located predominantly in the hippocampus. Spinal lesions consisted of focal degeneration, lymphocytic infiltrates, and mild neuronal necrosis (Fig. 5). Inflammatory lesions were more severe in the brain than in the spinal cord. In contrast, none of the mice inoculated with the Δ2A virus exhibited lesions in brain or spinal cord from day 1 to day 22 after inoculation.
V. EMCV1.26Δ2A is infectious but does not reach the CNS.
Throughout this study, we reported no clinical signs, no cell infiltration, and no tissue damage within the central nervous system in mice infected with the Δ2A virus, whereas all of these effects were detected in EMCV1.26-infected mice.
We wished to determine whether EMCV1.26Δ2A does not cause damage in the CNS because it is avirulent in these tissues, because it is unable to reach them, or because it is simply unable to replicate in vivo. Immunohistochemistry (Fig. 6) and real-time RT-PCR (Fig. 7) were performed for EMCV detection. At days 1, 2, 3, 4, 7, 9, and 22 after infection, hearts, brains, and thoracic and lumbar spinal cords were harvested from EMCV1.26- and EMCV1.26Δ2A-infected mice. In mice inoculated with wild-type virus, EMCV RNAs were detected in the heart as early as 1 day after infection in 2/5 mice, in 4/5 at day 2, and in all mice from day 4 after infection and onward (Fig. 7a and b). Despite the presence of EMCV in the heart, no clinical signs were observed before day 4 after infection.
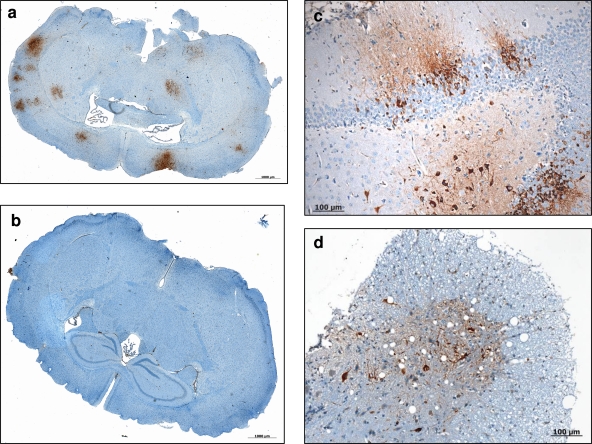
Fig. 6.
Immunohistochemistry of brains and spinal cords of C57BL/6 female mice i.p. injected with EMCV1.26 or EMCV1.26Δ2A. (a, b) Numerous foci of EMCV protein evidenced by immunohistochemistry in the brain of a mouse infected by EMCV1.26 (a), and the absence of any positive foci in the brain of a mouse infected by EMCV1.26Δ2A (b). (c, d) Higher magnification of neurons exhibiting positive staining with anti-EMCV antibody in the brain (c) and in the spinal cord (d) of a mouse infected by EMCV1.26. Hematoxylin counterstaining is shown.
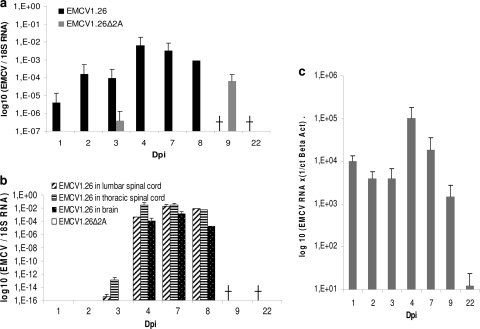
Fig. 7.
EMCV1.26Δ2A is infectious for mice but does not reach the central nervous system (CNS). EMCV and 18S real-time RT-PCR were performed on RNA extracted from mouse heart (a) and the CNS (b), including CNS tissues from the lumbar spinal cord (▨), thoracic spinal cord (▤), and brain (▩). Mice were inoculated with EMCV1.26 (black in panels a and b) or EMCV1.26Δ2A (gray in panel a; white in panel b). Five mice per indicated day and per virus were used for organ recovery. There were no more mice infected with EMCV1.26 at 9 and 22 dpi, as all were dead (†), and only one remained at day 8. (c) EMCV1.26Δ2A RNA is present in mouse heart but not in brain after inoculation of a high dose of virus. Twenty-one mice were inoculated with a high dose of EMCV1.26Δ2A (8 × 106 TCID50/mice), and 4 mice were inoculated with MEM as a negative control. Three mice per indicated day were used for organ recovery. EMCV and β-actin real-time RT-PCRs were performed on RNA extracted from mouse heart and brain. The graph represents the number of EMCV1.26Δ2A RNA copies in mouse heart at the indicated day postinoculation. No viral RNA was detected in any mouse brain.
In contrast, when inoculating the same amount of EMCV1.26Δ2A, we could detect only the Δ2A virus RNA in hearts of 1/5 mice at 3 days after infection. At 9 days after infection, 3/5 mice had significant amounts of virus RNA in the heart. Genomes were not detected afterwards for up to 22 days after infection. Inoculation of 200 times more viruses per mouse allowed detection of EMCV1.26Δ2A RNA in every mouse heart (Fig. 7c), but again, no clinical signs were detected. Detection of the EMCV1.26Δ2A genome long after inoculation (9 days after infection using low-dose inoculation and 22 days after high-dose inoculation) suggests that the Δ2A virus had replicated in vivo and was thus infectious for mice.
In the CNS, EMCV1.26 was first detected, by real-time RT-PCR, at 3 days after infection in the lumbar and thoracic spinal cords (in 1/5 mice at day 3) and at 4 days after infection in the brain, which corresponds to the onset of paralysis and death of mice (Fig. 7b). The EMCV antigen was readily detected by immunohistochemical analysis in neurons of the brain and spinal cord in association with lesions at 4 and 7 days after infection with EMCV1.26 (Fig. 6a, c, and d). The presence of virus in the CNS correlated with the appearance of lesions and clinical signs. In regard to the EMCV1.26Δ2A virus, however, virus was not detected in any of the CNS tissues tested either by real-time RT-PCR or by immunohistochemistry (Fig. 6b and 7b). Even after infection with a 200-times higher dose of virus, we could not detect EMCV RNA in mouse brain (Fig. 7c). Taken together, our results show that EMCV1.26Δ2A reached mouse heart but may be unable to reach the CNS. These findings could explain the nonpathogenic phenotype observed in mice with this mutant virus.
VI. Cell death and viral release of EMCV1.26Δ2A are substantially delayed in vitro.
In order to gain a better understanding of the mechanisms underlying the phenotype of the Δ2A virus, further in vitro studies were performed on BHK-21 cells. First, cell viability was examined at 10 and 24 h after infection at the indicated MOI (Fig. 8 a). All cells rapidly died upon infection with the wild-type EMCV1.26 virus in a time- and MOI-dependent manner. In contrast, at 10 h after infection, approximately 90% of cells infected with EMCV1.26Δ2A were still viable, irrespective of MOI (Fig. 8a, left). At 24 h (Fig. 8a, right), however, EMCV1.26Δ2A induced death in a substantial percentage of cells at high MOIs, suggesting that the virus conserved its ability to kill cells, albeit with lesser efficacy. Production of the Δ2A virus reached a plateau at 7 h (Fig. 3b). It appeared, however, that cells stayed alive even when the maximum production of new infectious particles was reached. Thus, EMCV1.26Δ2A may have a defect in killing cells and/or releasing new infectious particles.
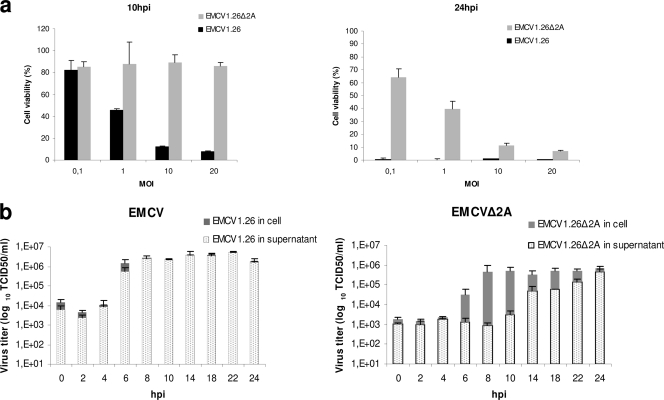
Fig. 8.
Cell death and viral release of EMCV1.26Δ2A are delayed in vitro. (a) Measure of cell viability, using the WST-1 reagent, 10 or 24 h after infection with EMCV1.26 (black) or EMCV1.26Δ2A (gray) at the indicated MOIs. (b) Kinetics of infectious particle release. Determination of titers of infectious particles (in TCID50/ml) in the intracellular compartment (gray) and supernatant (dots) over time for EMCV1.26 (left) and EMCV1.26Δ2A (right) after infection using an MOI of 10.
In order to examine viral release, the kinetics of viral production was reexamined for EMCV1.26 and EMCV1.26Δ2A, but this time, supernatant and cell lysate were titrated separately (Fig. 8b).
During EMCV1.26 infection, large amounts of viruses were detected in supernatant when the viral production had reached its maximum (at 6 h) (Fig. 8b, left), suggesting that newly produced viruses were only very briefly retained in the cells. Strikingly, EMCV1.26Δ2A tended to accumulate intracellularly (from 6 h onward) and to be progressively released (Fig. 8b, left), while EMCV1.26 release was fast and massive. These results reveal a defect in virus release correlating with a longer viability of cells infected with EMCV1.26Δ2A.
VII. EMCV1.26Δ2A-infected cells died by apoptosis.
Under a light microscope, cells infected with wild-type EMCV1.26 appeared “ghost-like” or to have lost intracellular content (characteristic of cytopathic cell death), while cells infected with the Δ2A virus rounded up and aggregated (data not shown). From the literature, it appeared that this second phenotype resembled apoptosis. To investigate whether cells infected by the EMCV1.26Δ2A virus died by apoptosis, cells were stained for cleaved caspase 3, a well-known marker of the last steps of the apoptotic process, and for chromatin condensation and nuclear fragmentation of DNA, a hallmark of apoptosis.
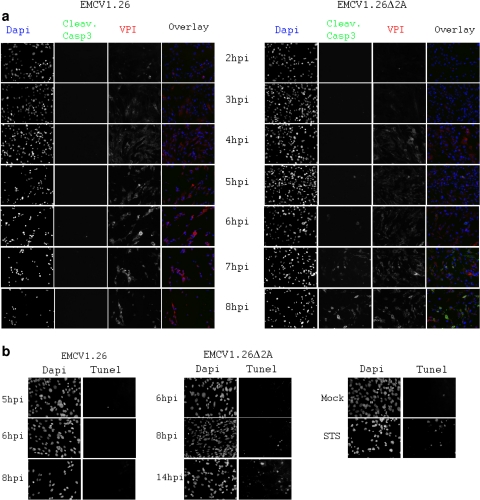
No signs of apoptosis could be detected in EMCV1.26-infected BHK-21 cells at 8 and 14 h after infection (Fig. 9 a) or at earlier time points (Fig. 10 a). This has also been confirmed using another specific apoptotic labeling system, TUNEL staining of cells (Fig. 10b). No positive TUNEL cells were noticed following EMCV1.26 infection. These results agree with observations made by Romanova et al. (41), whereby cells infected with EMCV do not undergo apoptosis and indeed become refractory to apoptotic signals.
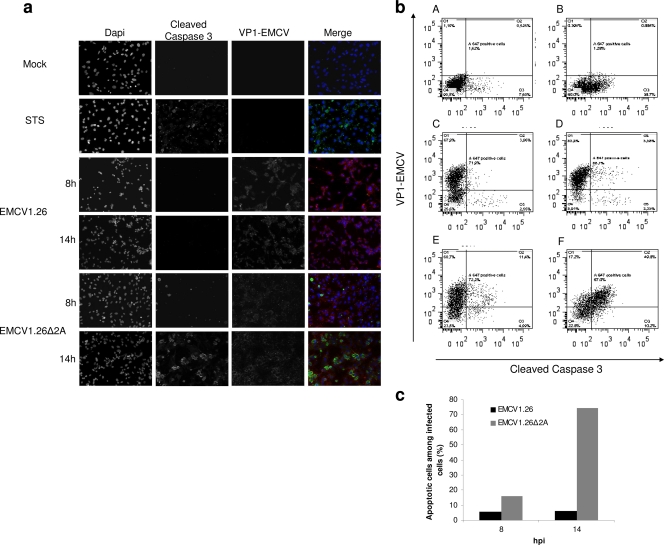
Fig. 9.
EMCV1.26Δ2A-infected cells die by apoptosis. (a) BHK-21 cells were mock infected, treated with STS for apoptosis induction, or infected at an MOI of 10 with EMCV1.26 or EMCV1.26Δ2A. Cells were fixed at the indicated time postinfection and immunostained with anti-EMCV VP1 followed by PE-labeled anti-mouse IgG (red), with anti-cleaved caspase 3 followed by Alexa 488-labeled anti-rabbit IgG (green), and with DAPI (blue). (b) Dot plots obtained by flow cytometry of BHK-21 cells that were untreated (A), treated with STS (B), and infected at an MOI of 5 with EMCV1.26 for 8 h (C) or 14 h (D) or with EMCV1.26Δ2A for 8 h (E) or 14 h (F). Cleaved caspase 3 (apoptosis) and EMCV VP1 (viral infection) are graphed on x and y axes, respectively. (c) Percentage of apoptotic cells among infected BHK-21 cells. EMCV1.26 (black); EMCV1.26Δ2A (gray).
Fig. 10.
EMCV1.26Δ2A-infected cells die by apoptosis, in contrast to EMCV1.26-infected cells. BHK-21 cells were infected at an MOI of 10 with EMCV1.26 or EMCV1.26Δ2A. (a, b) Cells were fixed at the indicated time postinfection and immunostained with anti-EMCV VP1 followed by Alexa 546-labeled anti-mouse IgG (red), with anti-cleaved caspase 3 followed by Alexa 488-labeled anti-rabbit IgG (green), and with DAPI (blue) (a) or labeled for DNA fragmentation using TUNEL staining (Promega) and DAPI (b).
In contrast, the Δ2A virus elicited a marked apoptosis in BHK-21 cells at 14 h, as judged by cell rounding, detection of cleaved caspase 3, chromatin condensation (Fig. 9a), and DNA fragmentation visualized by TUNEL staining (Fig. 10b). Moreover, nuclear fragmentation and formation of apoptotic bodies were detectable. At 8 h, minor signs of apoptosis were detected. While the infection itself may have triggered apoptosis, we could not exclude that apoptosis was an indirect effect, whereby uninfected cells underwent apoptosis in response to danger signals released by their infected neighbors. To determine the percentage of infected cells exhibiting apoptosis, we performed flow cytometry on cells infected with either wild-type or Δ2A virus. Cells were detached, permeabilized, and stained with anti-EMCV and anti-caspase 3 antibodies at the indicated times after infection.
Representative dot plots obtained for EMCV1.26/EMCV1.26Δ2A-infected cells and controls (STS treatment or mock) are represented in Fig. 9b. Regarding control samples, 8% of untreated cells were apoptotic, while approximately 40% of cells treated for 18 h with STS (a nonviral inducer of apoptosis) were. After inoculation with wild-type virus (MOI of 5) for 8 and 14 h, 72 and 89% of cells were infected (that is, positive for VP1), respectively, and the percentage of cells in which cleaved caspase 3 was detected was even lower than that of the untreated cells. Thus, EMCV1.26 infection did not lead to apoptosis, as we had suspected from immunostaining (Fig. 9a and 10). In contrast, 15% and 60% of cells inoculated with EMCV1.26Δ2A for 8 and 14 h, respectively, exhibited detectable levels of cleaved caspase 3. These results are summarized in Fig. 9c. Only 5 to 6.5% of cells infected with EMCV1.26 were undergoing apoptosis, while among cells infected with EMCV1.26Δ2A, 15.8% were apoptotic at 8 h, and this percentage rose to 74.2% at 14 h. Taken together, these results indicate that, in contrast to those infected with EMCV1.26, cells infected with EMCV1.26Δ2A died by apoptosis. This suggests that the 2A protein of EMCV virus is required for inhibition of apoptosis.
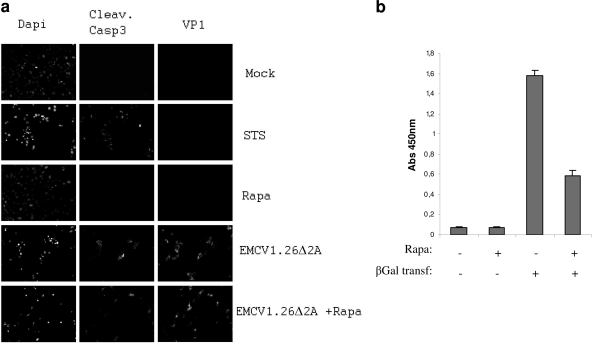
The 2A protein of EMCV has been shown to inhibit the cap-dependent translation in infected cells (20). To determine whether apoptosis occurs because of a defect of translation shutdown, we treated BHK-21 cells with rapamycin, a drug known to inhibit cap-dependent translation. We found that at 24 h posttreatment, rapamycin did not induce apoptosis (Fig. 11 a). We confirmed that rapamycin efficiently inhibits cap-dependent translation by transfecting the β-Gal reporter plasmid. Upon treatment, we measured a 60% decrease of the cap-dependent translation (Fig. 11b). Finally, we induced translation inhibition using rapamycin and infected cells with EMCV1.26Δ2A. Apoptotic signs were detected in treated as well as untreated infected cells (Fig. 11a). These results suggest that apoptosis occurring in EMCV1.26Δ2A-infected cells may not be due to an inefficient shutdown of cap-dependent translation.
Fig. 11.
EMCV1.26Δ2A-infected cells die by apoptosis even under inhibition of cap-dependent translation. (a) BHK-21 cells were treated with a 100 nM inhibitor of cap-dependent translation, rapamycin (Rapa), for 24 h before infection at an MOI of 10 with EMCV1.26Δ2A. Cells were fixed at 14 hpi and immunostained with anti-EMCV VP1 followed by Alexa 546-labeled anti-mouse IgG (VP1), with anti-cleaved caspase 3 followed by Alexa 488-labeled anti-rabbit IgG (Cleav. Casp3) and with DAPI. (b) The graph represents the inhibition of cap-dependent translation during rapamycin treatment. BHK-21 cells were treated or not with 100 nM rapamycin for 24 h before transfection of a reporter plasmid of the cap-dependent translation (pRSV-βGal). The β-galactosidase activity was measured. Untransfected cells treated or not with rapamycin were used as negative controls.
DISCUSSION
The EMCV is known to induce myocarditis, diabetes, and reproductive and nervous disorders (11, 28, 40, 48, 56). It is believed that disease is strain specific. Animal species, sex, and age are also known to be important factors for EMCV virulence. The two EMCV strains used in this study (B279/95 and 2887A/91, origin of EMCV1.26) induce different diseases in pigs (myocarditis and reproductive disorder, respectively [14]). Irrespective of the disease induced in pigs, both strains were highly neurovirulent in C57BL/6 mice (Fig. 1 and 4). We showed that both viruses induced hind-limb paralysis, encephalitis, and mouse death.
Histological studies showed lesions and EMCV antigen in the CNS of mice inoculated with EMCV1.26 (Fig. 5 and 6). Nervous lesions induced by EMCV1.26 consisted mainly of perivascular mononuclear infiltrate in the form of perivascular cuffs, multifocal neuronal necrosis, and diffuse gliosis. Cerebral lesions were located predominantly in the hippocampus, as previously observed for the EMCV 30/87 strain (29). Spinal cord lesions due to EMCV1.26 consisted of focal degeneration, lymphocytic infiltrates, and mild neuronal necrosis (Fig. 5). The spinal lesions we found were in agreement with those observed by Takeda et al. (48–50) for the EMCV-D strain. Inflammatory lesions were more severe in the brain than in the spinal cord, but high levels of EMCV1.26 antigen were also detected in the spinal cord (Fig. 6).
During EMCV infection, hind-limb paralysis and demyelinization have been proven to be partly due to the infiltration of macrophages (49, 50). Treatment of mice with antibodies raised against macrophages and CD4+ T lymphocytes reduced lesions in spinal cords and incidence of paralysis, indicating that macrophages and CD4+ T cells participate in CNS injury (45, 51). Also, Ano et al. (3) demonstrated that after intracranial inoculation of mice with EMCV, oxidative damage to neurons was caused by induction of microglial NADPH oxidase. We also observed cell infiltrations and tissue damage during EMCV infection (Fig. 5 and 6). These studies indicate that EMCV neurovirulence might be due to or at least is exacerbated by the inflammatory response. This response may not, however, be peculiar to development of neurovirulence, as it has also been described in diabetes (56).
The molecular determinants of EMCV virulence are not fully defined, and no data have been published regarding the molecular determinants of EMCV neurovirulence in mice. Here, we showed that B279/95p210, which is avirulent for pigs (13), remained neurovirulent for mice, although less so than the wild-type virus (Fig. 1). One of the isolated clones, B279/95p210C9, was totally avirulent for mice. This clone had two mutations in the L protein, three in VP2, and five in VP1. These mutations, however, were also found in some other clones that conserved neurovirulence for mice (data not shown). Interestingly, the C9 clone was the only clone to present a deletion in the 2A protein.
Introduction of this deletion into EMCV1.26, which induces neurological symptoms similar to those of B279/95 when inoculated in mice, yielded a virus (EMCV1.26Δ2A) (Fig. 2) that was attenuated in mice (Fig. 4). The deletion-bearing virus did not induce any clinical signs, even under high-dose inoculation (Fig. 7c), and no lesions were found in the CNS of inoculated mice (Fig. 5). Real-time RT-PCR performed on mouse heart showed that EMCV1.26Δ2A RNA was present in all mouse hearts when inoculated with a high dose of EMCV1.26Δ2A (Fig. 7c). These results indicate that the Δ2A virus was indeed infectious in vivo and able to disseminate at least in the heart.
Nevertheless, the EMCV1.26Δ2A virus was not found in the CNS, whether expression of viral RNA or protein was sought (Fig. 6b and 7b). Even when inoculating 200 times more virus, no viral RNA was found in mouse brain (Fig. 7c). We interpreted this observation as meaning that expression of EMCV1.26Δ2A in mouse heart did not suffice to induce clinical signs. For mice inoculated with the wild-type virus, clinical signs appeared concomitantly with the detection of virus in spinal cords and brain (Fig. 6 and 7a and b). These results suggest that EMCV1.26 had to reach the CNS to induce clinical signs and thus that EMCV1.26Δ2A may not be pathogenic because it did not reach the CNS.
The fact that EMCV1.26Δ2A virus did not reach the CNS could possibly be explained by the defect in virus release together with lower production of new infectious particles, as was observed in vitro (Fig. 3 and 8).
Studies implicating different types of viruses have demonstrated that defects in virus release in vitro give rise to in vivo attenuation (18, 30, 32, 53). For a rapidly lytic virus such as EMCV, it seems likely that impaired release would decrease propagation and hence contribute to its attenuated phenotype.
The small-plaque phenotype observed with EMCV1.26Δ2A (Fig. 3a) led us to presume that cell-to-cell propagation was hindered. Indeed, EMCV1.26Δ2A presented a defect in virus release coupled with prolonged cell viability during BHK-21 infection (Fig. 8). This prolongation in cell viability could hardly be attributed to the 1-h delay observed in the one-step growth curve of the deleted virus in comparison with that of the wild-type virus (Fig. 3b). Even if virus production had reached its maximum, virions seemed to remain trapped in the cell (Fig. 8). Indeed, these observations raise the remaining question of what exactly leads to cell lysis during cardiovirus infection.
The 1-h delay in production of new infectious particles observed for EMCV1.26Δ2A in Fig. 3 may be linked to less efficient cleavage of P1-1.26Δ2A, as Svitkin et al. (47) have observed using two EMCVs deleted in 2A (deletions from aa 5 to 125 and aa 39 to 97, which overlap our deletion). Indeed, higher accumulation of uncleaved P1-Δ2A might delay the capsid assembly and thus production of new infectious particles. The EMCV1.26Δ2A virus produced for this study has an extensively truncated 2A protein (28 aa in Δ2A, compared with 143 aa in the wild-type virus). From recent analyses of viruses bearing mutations in 2A that defined motifs within the 2A protein (20), it appears that the truncated 2A protein encoded by EMCV1.26Δ2A does not possess an NLS but conserves the terminal 18 aa with the eIF4E binding site and the NPG(P) sequence required for the first “cleavage.” Groppo et al. demonstrated that when the 2A NLS is disrupted the protein no longer localizes to the nucleus and that the inhibition of host cell translation is ineffective (20). This led us to presume that during EMCV1.26Δ2A infection, cellular cap-dependent translation may not be inhibited. This might also be a partial explanation of the 1-h delay of viral production (due to cap/IRES translation competition), but also it might help cells having a longer viability. Cell viability was evaluated by quantifying the total metabolic activity. At 10 hours postinfection (hpi), it might be too early to measure a real decrease in cell metabolism, even if the cell started the process of apoptosis (Fig. 8, 9, and 10). Moreover, for the viability observed at 24 h, possible arguments are that by using an MOI of 1, approximately 50% of cells are really infected, even for EMCV1.26 (Fig. 2b), and that EMCV1.26Δ2A takes more time to achieve one complete cycle, and thus, takes more time to spread at low MOIs.
The EMCV has been described mainly as a lytic virus that undergoes “necrotic” cell death, which is in agreement with our observations, whereby infection of BHK-21 cells with EMCV1.26 resulted in their cytopathic death. Unexpectedly, we found that BHK-21 cells infected by the Δ2A virus underwent cell death by a different pathway. We found that cells infected with EMCV1.26Δ2A died by apoptosis (Fig. 9 and 10) through caspase activation. Considering that EMCV1.26-infected cells did not undergo programmed cell death (Fig. 9 and 10), that EMCV has been shown to inhibit apoptosis (1, 41), and that cells infected with a virus deleted in 2A were apoptotic, it is likely that the EMCV 2A protein is in some way required to inhibit apoptosis. It is a possibility that apoptosis occurs due to the noninhibition of host cell protein synthesis by 2A. However, we present data showing that the inhibition of the cap-dependent translation did not avoid apoptosis in EMCV1.26Δ2A-infected cells (Fig. 11). These suggest that the 2A protein is required for inhibition of apoptosis, but it might not be due to its ability to inhibit cap-dependent translation.
Interestingly, it has been shown that EMCV is able to inhibit apoptosis even under an apoptosis inducer (1, 41), but use of a mengovirus (closely related to EMCV) with mutation in the L finger zing domain made the virus unable to inhibit apoptosis.
It is known for Theiler's murine encephalitis virus (TMEV) that its L protein induces apoptosis, while the L* protein inhibits it (17, 38). EMCV does not posses an L* protein, so the mechanisms that contribute to the regulation of apoptosis in EMCV-infected cells remain to be better characterized, especially how these two “security proteins,” L and 2A, may contribute to this phenomenon.
In vivo, lytic cell death, in comparison to apoptosis, gives rise to a greater inflammatory response (15, 19), which is supposed to exacerbate EMCV virulence, as discussed above. In addition, inhibition of apoptosis by NF-κB has been shown to be required for pathogenesis during EMCV infection in mice (44). Indeed, NF-κB is a transcription factor known to induce inflammatory responses and to inhibit apoptosis (23) and is activated during EMCV infection (42). Thus, inhibition of apoptosis as well as induction of a strong inflammatory response might also be important factors for EMCV virulence.
Of note, wild-type EMCV infection has been described to induce apoptosis in mice (26, 34, 37) and pigs (9). It is at present unclear whether apoptosis is a direct or indirect effect of viral infection. The possibility, as described by Buenz et al. (10) for TMEV infection in mice, that the majority of cells triggering apoptosis were uninfected cannot be ruled out. Whether induction of apoptosis during EMCV infection in vivo results from the virus itself or from the host response to viral infection is not yet understood.
However, alleviating the consequence of inflammation may not be sufficient to stop the pathological process. As mentioned earlier, lower virus production and delay in EMCV1.26Δ2A virus production, release, spreading, and cell death may facilitate development of an immune response capable of clearing the virus. Virus trapped in cells might be phagocytosed more easily, particularly if the cell is undergoing apoptosis (15).
In conclusion, the EMCV 2A protein is important for the virus to counteract the host defense, as Δ2A viruses do not inhibit cap-dependent translation (20), are unable to inhibit apoptosis in vitro, and are no longer pathogenic in vivo. The 2A protein thus well deserves to be considered a “viral security protein” (1).
Strikingly, deletion of 115 amino acids in the 2A protein of two different EMCV strains profoundly affected their virulence in vivo. In addition, EMCV1.26Δ2A-infected cells underwent apoptosis, unlike those infected with the wild-type virus. This strongly suggests that the 2A protein of EMCV is required to avoid apoptosis, which may be important for viral pathogenesis. This study suggests a link between viral release, antiapoptotic activity, and pathogenicity. Further investigations are, however, needed to elucidate how virus may manipulate cell death processes to improve viral dissemination.
ACKNOWLEDGMENTS
This research was funded by the European Commission (FP7 grant 226556). M.C. was supported by a fellowship from the ANSES.
We thank Jennifer Richardson for editing the English version of the manuscript and Alenxandra Desprat, Oceane Le Bidel, and Alain Bernier for their technical assistance.
We have no conflicts of interest to report.
Footnotes
Published ahead of print on 17 August 2011.
REFERENCES
- 1. Agol V. I., Gmyl A. P. 2010. Viral security proteins: counteracting host defences. Nat. Rev. Microbiol. 8:867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aminev A. G., Amineva S. P., Palmenberg A. C. 2003. Encephalomyocarditis viral protein 2A localizes to nucleoli and inhibits cap-dependent mRNA translation. Virus Res. 95:45–57 [DOI] [PubMed] [Google Scholar]
- 3. Ano Y., et al. 2010. Oxidative damage to neurons caused by the induction of microglial NADPH oxidase in encephalomyocarditis virus infection. Neurosci. Lett. 469:39–43 [DOI] [PubMed] [Google Scholar]
- 4. Autret A., et al. 2008. Early phosphatidylinositol 3-kinase/Akt pathway activation limits poliovirus-induced JNK-mediated cell death. J. Virol. 82:3796–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bae Y. S., Yoon J. W. 1993. Determination of diabetogenicity attributable to a single amino acid, Ala776, on the polyprotein of encephalomyocarditis virus. Diabetes 42:435–443 [DOI] [PubMed] [Google Scholar]
- 6. Bedard K. M., Semler B. L. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6:702–713 [DOI] [PubMed] [Google Scholar]
- 7. Beretta L., Svitkin Y. V., Sonenberg N. 1996. Rapamycin stimulates viral protein synthesis and augments the shutoff of host protein synthesis upon picornavirus infection. J. Virol. 70:8993–8996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Breiman A., et al. 2005. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon. J. Virol. 79:3969–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brewer L. A., et al. 2001. Porcine encephalomyocarditis virus persists in pig myocardium and infects human myocardial cells. J. Virol. 75:11621–11629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buenz E. J., et al. 2009. Apoptosis of hippocampal pyramidal neurons is virus independent in a mouse model of acute neurovirulent picornavirus infection. Am. J. Pathol. 175:668–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cerutis D. R., Bruner R. H., Thomas D. C., Giron D. J. 1989. Tropism and histopathology of the D, B, K, and MM variants of encephalomyocarditis virus. J. Med. Virol. 29:63–69 [DOI] [PubMed] [Google Scholar]
- 12. Cui T., Sankar S., Porter A. G. 1993. Binding of encephalomyocarditis virus RNA polymerase to the 3′-noncoding region of the viral RNA is specific and requires the 3′-poly(A) tail. J. Biol. Chem. 268:26093–26098 [PubMed] [Google Scholar]
- 13. Denis P., Koenen F. 2003. Molecular analysis of the capsid coding region of a virulent encephalomyocarditis virus isolate after serial cell passages and assessment of its virulence. Arch. Virol. 148:903–912 [DOI] [PubMed] [Google Scholar]
- 14. Denis P., et al. 2006. Genetic variability of encephalomyocarditis virus (EMCV) isolates. Vet. Microbiol. 113:1–12 [DOI] [PubMed] [Google Scholar]
- 15. Edinger A. L., Thompson C. B. 2004. Death by design: apoptosis, necrosis and autophagy. Curr. Opin. Cell Biol. 16:663–669 [DOI] [PubMed] [Google Scholar]
- 16. Esfandiarei M., et al. 2007. Coxsackievirus B3 activates nuclear factor kappa B transcription factor via a phosphatidylinositol-3 kinase/protein kinase B-dependent pathway to improve host cell viability. Cell. Microbiol. 9:2358–2371 [DOI] [PubMed] [Google Scholar]
- 17. Fan J., Son K. N., Arslan S. Y., Liang Z., Lipton H. L. 2009. Theiler's murine encephalomyelitis virus leader protein is the only nonstructural protein tested that induces apoptosis when transfected into mammalian cells. J. Virol. 83:6546–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fultz P. N., et al. 2001. In vivo attenuation of simian immunodeficiency virus by disruption of a tyrosine-dependent sorting signal in the envelope glycoprotein cytoplasmic tail. J. Virol. 75:278–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Galluzzi L., et al. 2010. Viral strategies for the evasion of immunogenic cell death. J. Intern. Med. 267:526–542 [DOI] [PubMed] [Google Scholar]
- 20. Groppo R., Brown B. A., Palmenberg A. C. 2011. Mutational analysis of the EMCV 2A protein identifies a nuclear localization signal and an eIF4E binding site. Virology 410:257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Groppo R., Palmenberg A. C. 2007. Cardiovirus 2A protein associates with 40S but not 80S ribosome subunits during infection. J. Virol. 81:13067–13074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hahn H., Palmenberg A. C. 2001. Deletion mapping of the encephalomyocarditis virus primary cleavage site. J. Virol. 75:7215–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hayden M. S., Ghosh S. 2004. Signaling to NF-kappaB. Genes Dev. 18:2195–2224 [DOI] [PubMed] [Google Scholar]
- 24. Jen G., Detjen B. M., Thach R. E. 1980. Shutoff of HeLa cell protein synthesis by encephalomyocarditis virus and poliovirus: a comparative study. J. Virol. 35:150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones P., et al. 2011. Encephalomyocarditis virus mortality in semi-wild bonobos (Pan panicus). J. Med. Primatol. 40:157–163 [DOI] [PubMed] [Google Scholar]
- 26. Kanda T., et al. 1999. Increased severity of viral myocarditis in mice lacking lymphocyte maturation. Int. J. Cardiol. 68:13–22 [DOI] [PubMed] [Google Scholar]
- 27. Kassimi L. B., Boutrouille A., Gonzague M., Mbanda A. L., Cruciere C. 2002. Nucleotide sequence and construction of an infectious cDNA clone of an EMCV strain isolated from aborted swine fetus. Virus Res. 83:71–87 [DOI] [PubMed] [Google Scholar]
- 28. Kim H. S., Christianson W. T., Joo H. S. 1989. Pathogenic properties of encephalomyocarditis virus isolates in swine fetuses. Arch. Virol. 109:51–57 [DOI] [PubMed] [Google Scholar]
- 29. LaRue R., et al. 2003. A wild-type porcine encephalomyocarditis virus containing a short poly(C) tract is pathogenic to mice, pigs, and cynomolgus macaques. J. Virol. 77:9136–9146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Low A., et al. 2007. Mutation in the glycosylated gag protein of murine leukemia virus results in reduced in vivo infectivity and a novel defect in viral budding or release. J. Virol. 81:3685–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu Q. 2005. Seamless cloning and gene fusion. Trends Biotechnol. 23:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marcucci K. T., Martina Y., Harrison F., Wilson C. A., Salomon D. R. 2008. Functional hierarchy of two L domains in porcine endogenous retrovirus (PERV) that influence release and infectivity. Virology 375:637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Medvedkina O. A., Scarlat I. V., Kalinina N. O., Agol V. I. 1974. Virus-specific proteins associated with ribosomes of Krebs-II cells infected with encephalomyocarditis virus. FEBS Lett. 39:4–8 [DOI] [PubMed] [Google Scholar]
- 34. Nakayama Y., Su W., Ohguchi A., Nakayama H., Doi K. 2004. Experimental encephalomyocarditis virus infection in pregnant mice. Exp. Mol. Pathol. 77:133–137 [DOI] [PubMed] [Google Scholar]
- 35. Neznanov N., et al. 2001. Poliovirus protein 3A inhibits tumor necrosis factor (TNF)-induced apoptosis by eliminating the TNF receptor from the cell surface. J. Virol. 75:10409–10420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oberste M. S., et al. 2009. Human febrile illness caused by encephalomyocarditis virus infection, Peru. Emerg. Infect. Dis. 15:640–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ohguchi A., et al. 2006. Encepalomyocarditis virus-induced apoptosis and ultrastructural changes in the lacrimal and parotid glands of mice. Exp. Mol. Pathol. 80:201–207 [DOI] [PubMed] [Google Scholar]
- 38. Okuwa T., Taniura N., Saito M., Himeda T., Ohara Y. 2010. Opposite effects of two nonstructural proteins of Theiler's murine encephalomyelitis virus regulates apoptotic cell death in BHK-21 cells. Microbiol. Immunol. 54:639–643 [DOI] [PubMed] [Google Scholar]
- 39. Palmenberg A. C., et al. 1984. The nucleotide and deduced amino acid sequences of the encephalomyocarditis viral polyprotein coding region. Nucleic Acids Res. 12:2969–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Psalla D., et al. 2006. Pathogenesis of experimental encephalomyocarditis: a histopathological, immunohistochemical and virological study in mice. J. Comp. Pathol. 135:142–145 [DOI] [PubMed] [Google Scholar]
- 41. Romanova L. I., et al. 2009. Antiapoptotic activity of the cardiovirus leader protein, a viral “security” protein. J. Virol. 83:7273–7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roos F. C., et al. 2010. Oncolytic targeting of renal cell carcinoma via encephalomyocarditis virus. EMBO Mol. Med. 2:275–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salako M. A., Carter M. J., Kass G. E. 2006. Coxsackievirus protein 2BC blocks host cell apoptosis by inhibiting caspase-3. J. Biol. Chem. 281:16296–16304 [DOI] [PubMed] [Google Scholar]
- 44. Schwarz E. M., et al. 1998. NF-kappaB-mediated inhibition of apoptosis is required for encephalomyocarditis virus virulence: a mechanism of resistance in p50 knockout mice. J. Virol. 72:5654–5660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sriram S., Topham D. J., Huang S. K., Rodriguez M. 1989. Treatment of encephalomyocarditis virus-induced central nervous system demyelination with monoclonal anti-T-cell antibodies. J. Virol. 63:4242–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stellmann C., Bornarel P. 1971. Calculation tables of D 50 titres of viral suspensions and their accuracy. Ann. Inst. Pasteur (Paris) 121:825–833 [PubMed] [Google Scholar]
- 47. Svitkin Y. V., Hahn H., Gingras A. C., Palmenberg A. C., Sonenberg N. 1998. Rapamycin and wortmannin enhance replication of a defective encephalomyocarditis virus. J. Virol. 72:5811–5819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takeda M., Hirasawa K., Doi K. 1991. Lesions in the central nervous system of DBA/2 mice infected with the D variant of encephalomyocarditis virus (EMC-D). J. Vet. Med. Sci. 53:1013–1017 [DOI] [PubMed] [Google Scholar]
- 49. Takeda M., Itagaki S., Doi K. 1993. Biphasic disease of central nervous system induced in DBA/2 mice by the D variant of encephalomyocarditis virus (EMC-D). Int. J. Exp. Pathol. 74:493–499 [PMC free article] [PubMed] [Google Scholar]
- 50. Takeda M., et al. 1995. Distribution of viral RNA in the spinal cord of DBA/2 mice developing biphasic paralysis following infection with the D variant of encephalomyocarditis virus (EMC-D). Int. J. Exp. Pathol. 76:441–447 [PMC free article] [PubMed] [Google Scholar]
- 51. Takeda M., Ohtsuka R., Nakayama Y., Doi K. 2004. The role of CD4(+) T cells in biphasic hind limb paralysis induced by the D variant of encephalomyocarditis virus (EMC-D) in DBA/2 mice. Exp. Anim. 53:31–35 [DOI] [PubMed] [Google Scholar]
- 52. Tanaka N., et al. 1998. Type I interferons are essential mediators of apoptotic death in virally infected cells. Genes Cells 3:29–37 [DOI] [PubMed] [Google Scholar]
- 53. Vanderplasschen A., Hollinshead M., Smith G. L. 1997. Antibodies against vaccinia virus do not neutralize extracellular enveloped virus but prevent virus release from infected cells and comet formation. J. Gen. Virol. 78(Pt. 8):2041–2048 [DOI] [PubMed] [Google Scholar]
- 54. Vlemmas J., et al. 2000. Immunohistochemical detection of encephalomyocarditis virus (EMCV) antigen in the heart of experimentally infected piglets. J. Comp. Pathol. 122:235–240 [DOI] [PubMed] [Google Scholar]
- 55. Yeung M. C., Chang D. L., Camantigue R. E., Lau A. S. 1999. Inhibitory role of the host apoptogenic gene PKR in the establishment of persistent infection by encephalomyocarditis virus in U937 cells. Proc. Natl. Acad. Sci. U. S. A. 96:11860–11865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yoon J. W., Jun H. S. 2006. Viruses cause type 1 diabetes in animals. Ann. N. Y. Acad. Sci. 1079:138–146 [DOI] [PubMed] [Google Scholar]