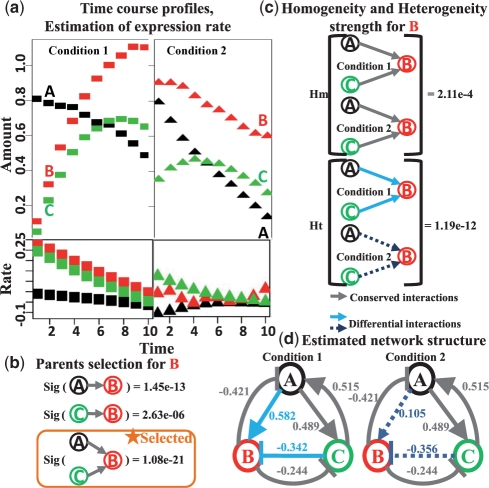
Fig. 2.
Overview of the comparative dynamical system modeling (CDSM) framework. Variables A, B and C represent three interacting molecules. (a) The input to CDSM is time courses of the three molecules under two conditions. Each series of time course data contains 10 time points. The expression rate for each variable is estimated from the time courses by pspline. (b) With variable B as an example, its parents are selected to be A and C, which represent the most significant total regulation strength among all possible interactions. (c) Homogeneity and heterogeneity across two conditions are calculated for the selected parent combination. The interaction for B showed significant heterogeneity of P-value 1.19e-12. Thus, we considered the interaction for B differential across the two conditions. (d) After applying CDSM on all three variables, we identified two conserved interactions targeting A and C and one differential interaction targeting B.