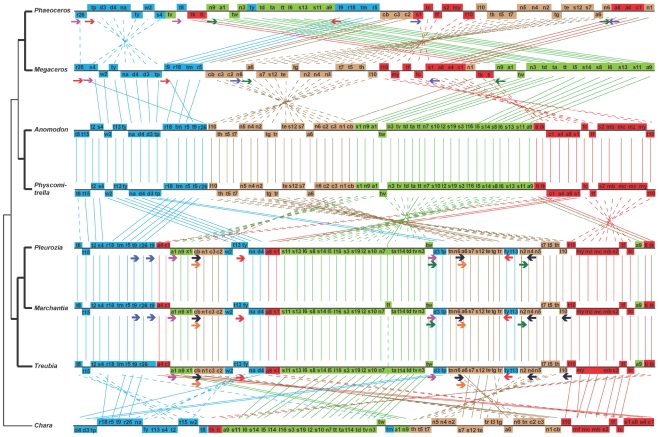
Figure 3. Gene order comparison among mitochondrial genomes of Chara vulgaris, Treubia lacunosa, Marchantia polymorpha, Pleurozia purpurea, Physcomitrella patens, Anomodon rugelii, Phaeoceros laevis, and Megaceros aenigmaticus.
Species are arranged according to the organismal phylogeny [39], [40], [42], [81] except that positions of the two hornworts are reversed as the Megaceros gene order more likely represents the ancestral condition according to a parsimony criterion (a supplementary figure (Fig. S1) is presented in which the two hornworts are placed in their correct organismal phylogeny positions). Solid lines connect orthologous genes between species with the same orientation, whereas dashed lines connect those with the reversed orientation. Repeat sequences are color-coded: in liverworts, RepA – black, RepB – green, RepB2 – purple, RepC – red, RepD – blue, and RepE – orange; in hornworts, RepA – red, RepB – blue (responsible for the inversion between Megaceros and Phaeoceros), RepC – green (this class of repeats was not annotated in either hornwort due to their length of <100 bp, inverted in Megaceros but direct in Phaeoceros), and RepD – purple.