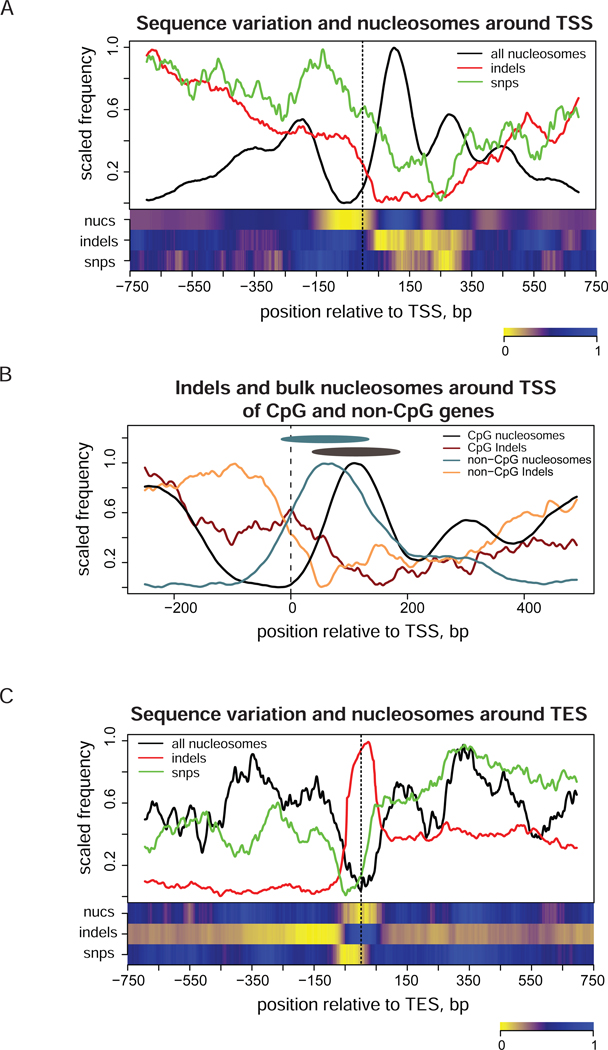
Figure 4.
Distribution of indels (red), SNPs (green), and stable nucleosome positions (black) around TSS and TES of human genes. Profiles were calculated as described in Methods. Heatmaps shown at the bottom panels represent de-trended profiles where large-scale variations were removed. (A) Profiles around TSS (position zero). The combined nucleosome set (‘all nucleosomes’) was used to produce this plot. Genes were oriented in the direction of transcription in such a way that the up-stream region is shown on the left and the downstream region is shown on the right of TSS. (B) Profiles shown separately for the frequencies of indels (dark red and orange lines) and bulk nucleosomes (black and cyan lines) for the subsets of genes associated and not associated with CpG islands at TSS. Black and cyan ovals represent nucleosomes at position +1 in CpG and non-CpG genes and are shown for a nucleosome size reference. Coordinates of CpG islands were taken from USCS genome browser annotation34. (C) Profiles computed around TES (position zero) for all genes. The combined nucleosome set was used.