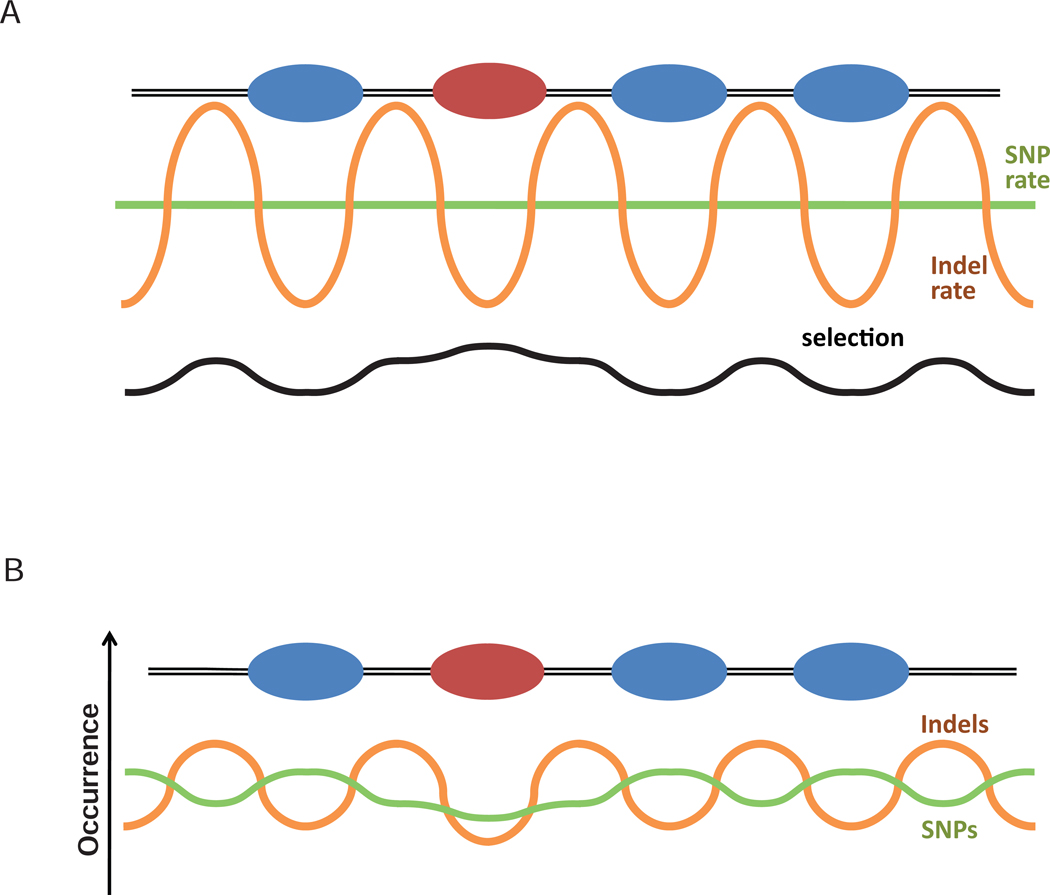
Figure 6.
Interplay of chromatin-mediated mutation bias and selection can shape sequence variation profile (cf. to schematic illustration in Ref. 27). (A) Bulk and epigenetically modified nucleosomes are represented with blue and red ovals. Green and orange lines represent mutation rate of SNPs and indels respectively, and black line represents selection pressure acting on the DNA sequence. (B) The significant difference in the indel rate inside and outside nucleosomes mainly determines the indel density profile observed in the genome (orange), while SNP density profile (green) is mainly affected by selection. Our results do not exclude the possibility that natural selection can affect the distribution of indels and that alteration of the mutation rate affects the distribution of SNPs. Rather, they indicate that these mechanisms are not the major factors shaping the resulting profiles.