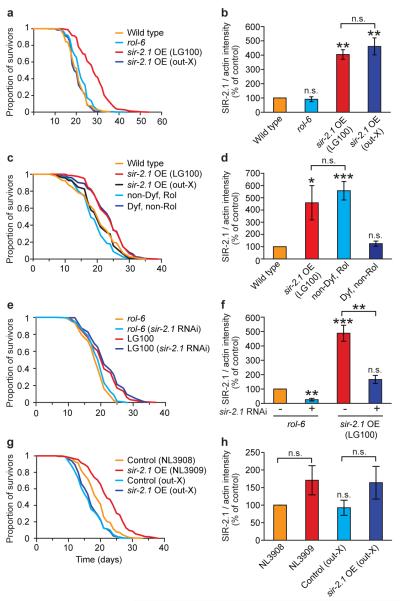
Figure 1. C. elegans: Longevity of LG100 and NL3909 is not attributable to sir-2.1 over-expression.
a, b. Outcrossing of LG100 removes life extension without affecting SIR-2.1 protein levels. Data in b derived from Western blots (mean of three trials each using an independent protein preparation). A representative Western blot is shown in Fig S1a. qRT-PCR showed that sir-2.1 mRNA is also elevated in both strains (data not shown). c, LG100-derived Dyf, non-Rol segregant lines are long-lived while non-Dyf, Rol lines are not. d. Non-Dyf Rol segregant lines have elevated SIR-2.1 levels, while Dyf, non-Rol lines do not. e, f. sir-2.1 RNAi does not suppress LG100 longevity, but reduces SIR-2 protein levels. g, h. Outcrossing of NL3909 removes life extension without affecting SIR-2.1 protein levels. See Tables S1, S3, S4 and S5 for lifespan statistics for a, c, e and g, respectively. Error bars, S.E.M.. *0.01< P < 0.05; ** 0.001 < P < 0.01; ***P < 0.001, n.s., not significant; Student’s t test (two tailed). One remaining possibility is that the outcrossed sir-2.1 strains both contain second site mutations that suppress longevity effects. However, daf-2 RNAi strongly induced longevity in both (data not shown), arguing against the presence of a general suppressor of longevity in each case.