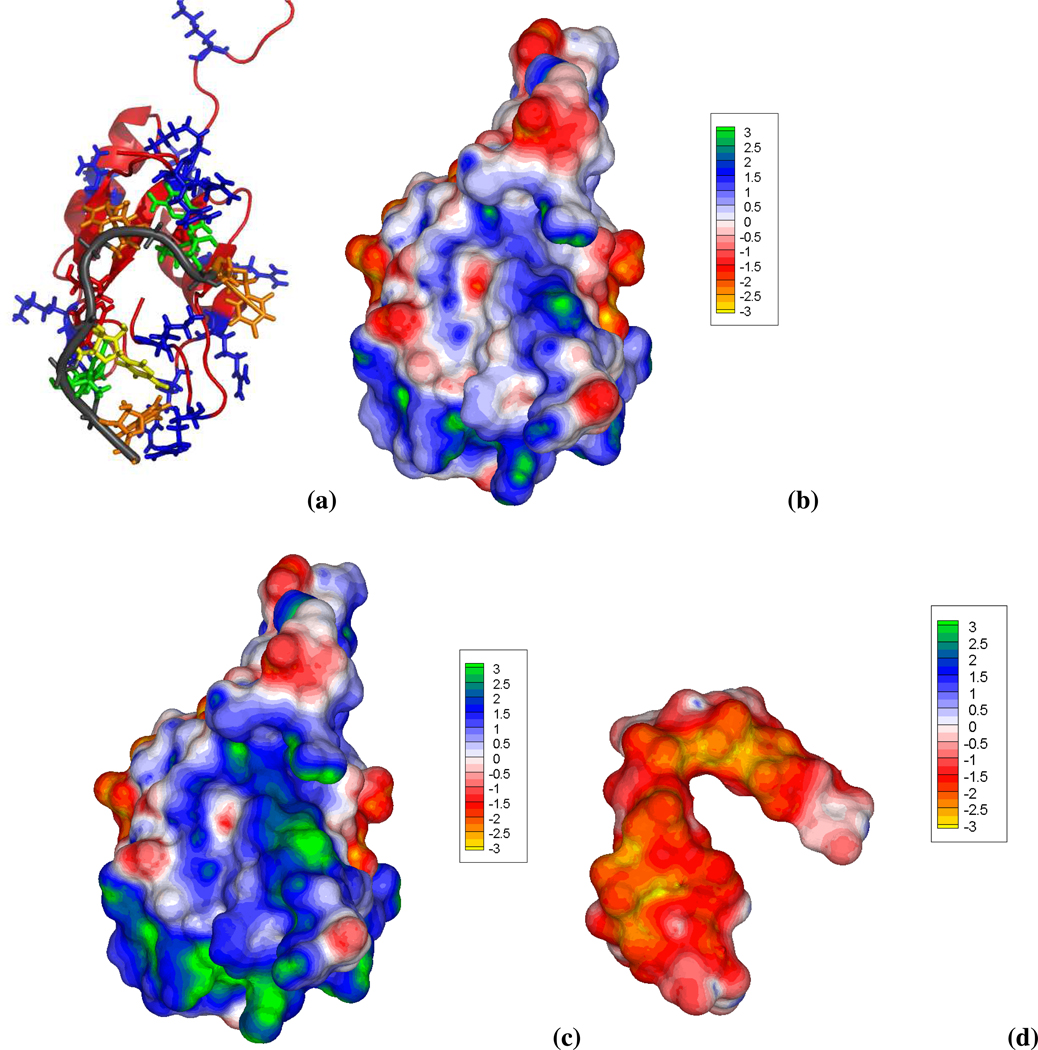
Figure 7.
Different depictions of the NMR structure of RNA binding domain (RBD) of Fox-1 in complex with the single-stranded UGCAUGU RNA element (PDB id: 2err, model 1). (a) RNA phosphate backbone depicted by the dark gray ribbon and the different bases colored as: adenine= red, uracil=orange, guanine=green and cytosine=yellow. The protein peptide backbone adopts an orange ribbon representation with positively and negatively charged side chains shown as blue and red sticks, respectively. This view emphasizes the clustering of various cationic protein residues at the RNA binding interface. (b) Surface electrostatic potential (in kcal/mol/e) and overall shape of the RBD of Fox-1. A well defined and concave region of positive potential, generated by the cationic protein residues and traced by the bent RNA structure, is clearly shown when the nonlinear PBE solution is employed. This cationic protein (net charge=+3e) follows the electrostatic pattern of other RNA binding proteins that have a distinct positive potential patch on their binding interface78. Thus, the RNA fills most of concave blue/green protein surface to which it is complementary in both shape and electrostatic potential. (c) Same view and color map as (b), but using the linear PBE. The positive electrostatic potential is now overestimated and the positive region much broader than in (b). (d) Electrostatic potential of the single-stranded bent and overall negatively charged RNA structure. As expected an overall negative potential covers most of the RNA surface due to the presence of the anionic phosphate groups.