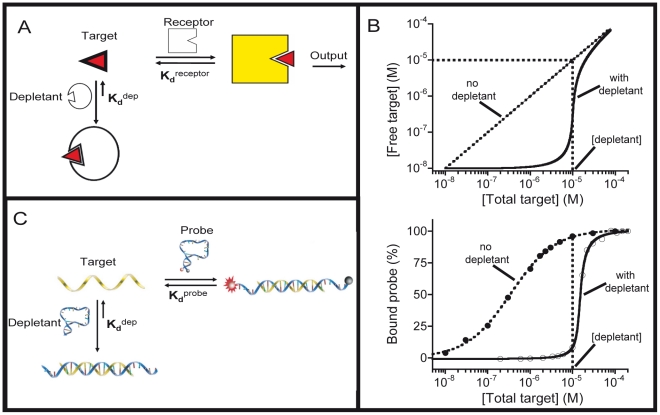
Figure 1. In vitro recreation of the sequestration mechanism.
Here we recapitulate in vitro the sequestration mechanism employed by nature to produce ultrasensitive outputs (steep input/output functions) in a number genetic regulatory networks. (a) In the sequestration mechanism low concentrations of a target molecule are “depleted” by binding to a high affinity non-signaling receptor that acts as a “sink” (the depletant). (b) Top, when the total target concentration surpasses the concentration of the depletant (the sink is saturated), a threshold response is achieved in which, upon the addition of any further target, the relative concentration of free target rises rapidly. Bottom, this threshold effect generates a “pseudo-cooperative” dose-response curve in which probe occupancy, and thus the output signal, arises much more rapidly than would occur in the absence of a depletant. (c) As a test bed to recapitulate and exploit this mechanism we have employed DNA molecular beacons, widely used probes for the detection of specific oligonucleotide sequences [29]. Consisting of a stem-loop DNA modified with a fluorophore/quencher pair the affinities of molecular beacons can be tuned by changing the stability of their stems [33], thus rendering it easy to generate depletant (unlabeled molecular beacons) with almost any arbitrary affinity.