Abstract
Abstract
Background
As scientists continue to pursue various 'omics-based research, there is a need for high quality data for the most fundamental 'omics of all: genomics. The bacterium Paenibacillus larvae is the causative agent of the honey bee disease American foulbrood. If untreated, it can lead to the demise of an entire hive; the highly social nature of bees also leads to easy disease spread, between both individuals and colonies. Biologists have studied this organism since the early 1900s, and a century later, the molecular mechanism of infection remains elusive. Transcriptomics and proteomics, because of their ability to analyze multiple genes and proteins in a high-throughput manner, may be very helpful to its study. However, the power of these methodologies is severely limited without a complete genome; we undertake to address that deficiency here.
Results
We used the Illumina GAIIx platform and conventional Sanger sequencing to generate a 182-fold sequence coverage of the P. larvae genome, and assembled the data using ABySS into a total of 388 contigs spanning 4.5 Mbp. Comparative genomics analysis against fully-sequenced soil bacteria P. JDR2 and P. vortex showed that regions of poor conservation may contain putative virulence factors. We used GLIMMER to predict 3568 gene models, and named them based on homology revealed by BLAST searches; proteases, hemolytic factors, toxins, and antibiotic resistance enzymes were identified in this way. Finally, mass spectrometry was used to provide experimental evidence that at least 35% of the genes are expressed at the protein level.
Conclusions
This update on the genome of P. larvae and annotation represents an immense advancement from what we had previously known about this species. We provide here a reliable resource that can be used to elucidate the mechanism of infection, and by extension, more effective methods to control and cure this widespread honey bee disease.
Background
Paenibacillus larvae is a spore-forming, Gram-positive bacterium, studied for the past century due to its ability to cause American foulbrood (AFB), a larval disease of honey bees [1]. The host is most vulnerable during an approximately 48-h window in the life cycle - the early larval stage - where arguably an undeveloped immune system and/or a lack of energy stores result in death. During this period, the oral LD50 ingestion is 8.49 spores [2]; death occurs due to systemic infection after the germinated bacterial spores proliferate in the midgut and then breach the midgut epithelium via a paracellular route [3]. The antibiotics oxytetracycline and tylosin are used both prophylactically and to treat symptoms; however, widespread drug resistance is evident [4] and their registered use is being withdrawn in many countries since residues can show up in honey. Even in susceptible isolates though, it is extremely difficult to completely eliminate from a hive, so without definitive knowledge of the molecular mechanism of pathogenesis, the design of specific treatments is significantly hindered. In 2006, a draft of the P. larvae genome was published at an estimated 5-6x coverage [5]; here we extend this coverage and further annotate the genome sequence with a combination of bioinformatics and proteomics.
Results and Discussion
Assembly of the P. larvae genome
Using the Illumina GAII platform and the ABySS assembler, together with the previously collected Sanger reads [5]; we achieved 182x coverage of the P. larvae genome and generated an initial assembly with a contig N50 of 49.6 kb. This statistic describes the contiguity of an assembly, and denotes that 50% of the reconstructed genome is contained in contigs equal to or larger than the given value. Contigs were joined into scaffolds using ABySS and Anchor (version 0.2.7, http://www.bcgsc.ca/platform/bioinfo/software/anchor/releases/0.2.7) (Docking et al., in preparation) to achieve a scaffold N50 of 83.2 kb. The largest contig size was 261,601 base pairs. There were 184 contigs with a size less than 1 kb, 136 contigs with length between 1 kb and 10 kb, 57 contigs with length between 10 kb and 100 kb and 11 contigs larger than 100 kb. The total length of the assembled contigs was 4,505,771 base pairs. Assembly statistics are listed in Table 1.
Table 1.
Summary of the P. larvae genome assembly.
| Read pairs | 8,212,402 |
|---|---|
| Read length | 50 bp |
| Fold coverage | 182x |
| Contig N50 | 49.6 kb |
| Scaffold N50 | 83.2 kb |
| Number of contigs | 388 |
| Total length of assembled contigs | 4,505,771 bp |
| Average GC content per contig | 44.04% |
| Mean contig size | 11.6 kb |
| Largest contig | 261,601 bp |
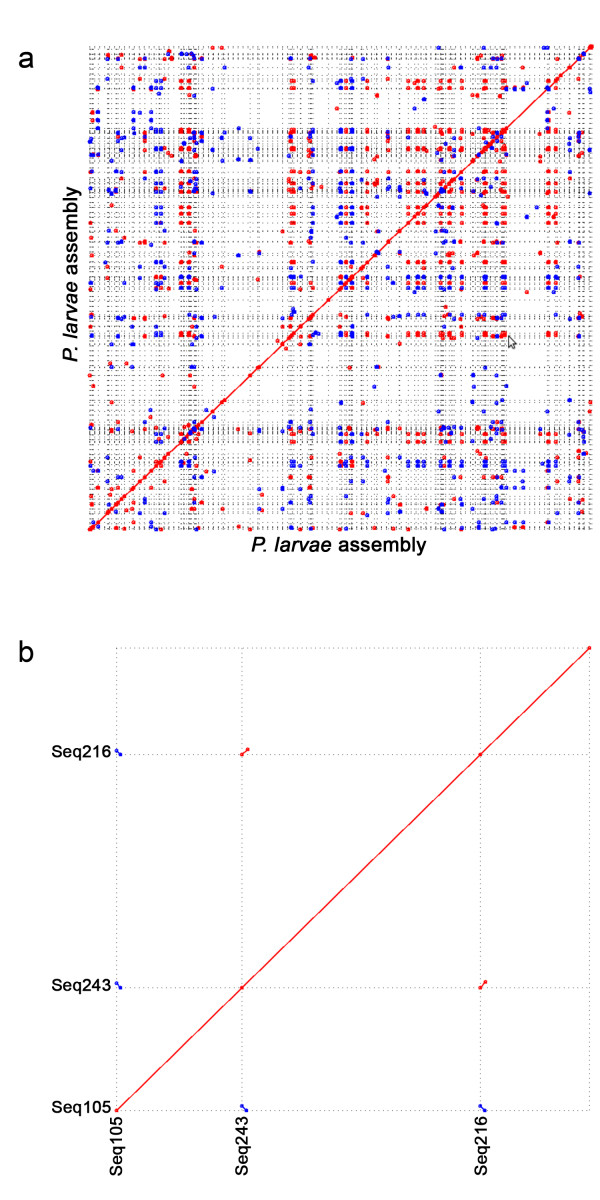
This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession ADZY00000000, with the latest version being ADZY02000000. Genome annotation and downstream analysis described below is based on the first version, ADZY01000000. Despite high sequence coverage, the assembly was relatively fragmented (Figure 1a). This fragmentation appears to be due primarily to long genomic repeats that could not be bridged by our sequencing strategy, as indicated by a preponderance of repetitive sequence occurring at contig ends (example shown in Figure 1b). Many of these repeats are similar to known bacterial insertion sequences and exceed 1 kb in length (data not shown). As a result, the majority of contigs were 1-10 kb in length.
Figure 1.
Self-alignment of 353 P. larvae contigs of assembly ADZY01000000.(a) MUMmer-generated graph of the P. larvae contigs aligned with themselves under default parameters. Red dots indicate sense matches and blue dots represent antisense matches. (b) Enlarged view of a sample region to demonstrate the common occurrence of alignments at the ends of contigs.
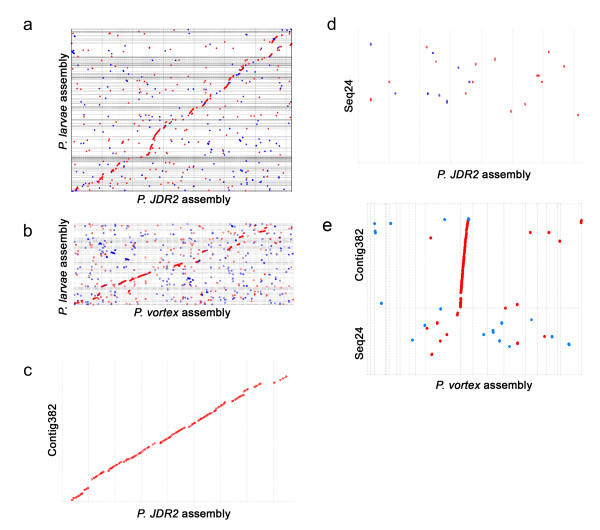
Given the fragmented nature of the assembly and the potential contribution of transposable elements to chromosome re-arrangements, we used tBLASTx and MUMmer to investigate the level of synteny between P. larvae and the congeneric soil bacterium P. JDR2. Although the completely sequenced P. JDR2 genome is 7.2 Mbp, substantially larger than our assembly (4.4 Mbp), the species share ~93% identity at 16S ribosomal loci. Figure 2a shows that, despite the difference in assembly size, gene-level synteny is generally conserved with P. JDR2 across the entirety of many P. larvae contigs. Some regions have excellent conservation (example in Figure 2b). This suggests that P. JDR2 can be a useful reference for comparative genomics and to order P. larvae contigs for genome finishing. Divergent regions between the two genomes are also of interest because they may harbor species-specific genes that are ecologically important in soil and beehive environments, respectively. For example, contig Seq24, which lacks synteny with P. JDR-2 (Figure 2c), contains two gene regions not present elsewhere in P. JDR-2 that are potential virulence factors (Table 2). Interesting, the same contig was also poorly conserved in another bacterium P. vortex (Figure 2d), thereby strengthening the claim that genes in this contig are unique to P. larvae. P. vortex is a pattern-forming soil bacterium with a 6.4 Mbp genome [6], so as expected, its overall level of synteny with P. larvae is quite high (Figure 2e), though slightly lower than observed when compared against P. JDR-2.
Figure 2.
Comparative genomics for aligning P. larvae contigs.MUMmer-generated dot-plot showing P. larvae contigs (y-axis) aligned with the fully-sequenced genomes of (a) P. JDR2 and (b) P. vortex (x-axes). Proteins sequences were used for matching. Red dots indicate same-direction matches and blue dots represent antisense matches. Magnified views of regions from (a) exemplify P. larvae contigs that are (c) highly conserved and (d) poorly conserved with P. JDR2; the same contigs are also compared in P. vortex (e).
Table 2.
Potential P. larvae virulence factors in genomic regions not found in P. JDR2.
| ORF | Strand | Start | Stop | Best match E-value | Best match taxon | Best match accession | Description |
|---|---|---|---|---|---|---|---|
| EFX43964 | minus | 1200 | 2189 | 1.00E-024 | Bacillus subtilis | NP_389716.2 | plipastatin synthetase |
| EFX43965 | minus | 2207 | 7786 | 1.00E-110 | Bacillus subtilis | NP_389598.3 | polyketide synthase of type I |
| EFX43966 | minus | 7790 | 12370 | 1.00E-107 | Streptococcus mutans | BAH88686.1 | NcpA protein |
| EFX43967 | minus | 12556 | 15906 | 0 | Bacillus subtilis | NP_389714.1 | plipastatin synthetase |
| EFX43968 | minus | 15902 | 19897 | 0 | Bacillus subtilis | NP_388230.2 | surfactin synthetase |
| EFX43969 | minus | 19920 | 25211 | 0 | Bacillus subtilis | NP_389714.1 | plipastatin synthetase |
| EFX43970 | minus | 25232 | 34267 | 0 | Bacillus subtilis | NP_388230.2 | surfactin synthetase |
| EFX43971 | minus | 34309 | 42072 | 0 | Bacillus subtilis | NP_389598.3 | polyketide synthase of type I |
| EFX44032 | minus | 110449 | 111363 | 3.00E-090 | Bacillus subtilis | NP_388265.1 | putative iron-siderophore ABC transporter (binding lipoprotein) |
| EFX44033 | minus | 111493 | 112248 | 1.00E-097 | Bacillus subtilis | NP_388264.1 | putative iron-siderophore ABC transporter (ATP-binding protein) |
| EFX44034 | minus | 112245 | 113180 | 9.00E-094 | Bacillus subtilis | NP_388263.1 | putative iron-siderophore ABC transporter (permease) |
| EFX44035 | minus | 113188 | 114057 | 8.00E-095 | Bacillus subtilis | NP_388262.1 | putative iron-siderophore ABC transporter (permease) |
Within the regions unique to P. larvae, one includes several open reading frames (ORFs) with strong homology to the synthetases of the antibiotic plipastatin, which inhibits phospholipase A2 [7] and surfactin [8], which possess hemolytic activity [9]. The region also contains putative type I polyketide synthetases whose products are secondary metabolites, some of which have antibiotic, immunosuppresant, or toxic effects [10]. The second region encodes homologs of a putative iron-siderophore ABC transporter, which enables iron uptake from the medium, an important process for many bacterial infections [11].
Annotation of putative P. larvae proteins
Using GLIMMER to predict genes from the P. larvae assembly and BLAST [12] searches against Bacillus or Streptococcus to provide annotation information (Additional File 1), we identified 3,568 gene models (final list in Additional File 2). Using shotgun proteomics methods to analyze P. larvae lysates, we identified 1262 proteins (1% false discovery rate), thus confirming the expression of 35% of the predicted proteins. Details regarding the identities of these proteins can be found in Additional File 3. As a major aim of this project is to find potential virulent factors and unique genes for this niche-specific bacterium, we describe below the genome content of P. larvae for several pathways considered to be important in this regard. The putative functions of the proteins were predicted by searching for matches against the Conserved Domain Database (CDD), and the complete results are tabulated in Additional File 4.
Flagellar system
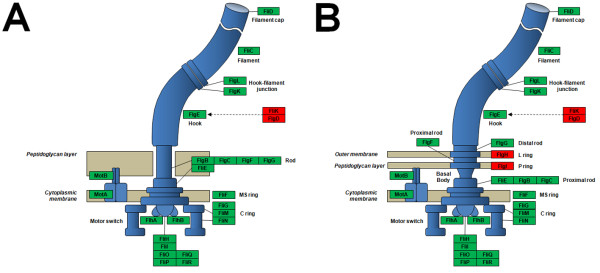
P. larvae is a flagellated bacterium and 41 genes associated with this system are detected in this assembly (Table 3), based on comparisons to two other fully-sequenced organisms. This group includes motor/switch, flagellar rings, rod hook and filament proteins, along with proteins involved in regulation and as chaperones. When compared against the Bacillus subtilis (Figure 3a, based on [13]) and Escherichia coli (Figure 3b, based on [14]) flagella, P. larvae has orthologs for nearly all of the genes. Those that are missing, such as the L and P ring proteins FlgH and FlgI in the outer membrane of the Gram-negative E. coli, are not necessary for the Gram-positive P. larvae since it only has one membrane [15]. However, it appears to be missing some players needed for assembly of the flagellar hook, a structure that acts both as a joint and motor for each individual flagellum. The hook itself, made of monomers, requires the monomer scaffolding protein FlgD [16]; this was not found in the P. larvae genome. Also missing is FliK, which acts essentially as a checkpoint for the flagellum's correct length prior to export. This may mean that this species has evolved ways to proceed with hook assembly despite their omission, or the enzymes were simply not detected by our current methods; as with many negative results, however, our inability to detect such genes does not imply P. larvae is completely incapable of quality control functions. Besides the flagellar structure, another important aspect is the control of its movement. Directionality is largely dictated by the Che gene family and methyl-accepting chemotaxis proteins [17], which can also be found in the P. larvae genome. The base of the flagellum consists of assembly and regulatory components, which are made of Fli and Mot gene families in E. coli [18]. Again, orthologs for most of these can be seen in P. larvae.
Table 3.
Putative P. larvae flagellar proteins
| Gene | Protein, functional information from CDD | P. larvae protein accession | |
|---|---|---|---|
| Two component system chemotaxis proteins | cheA | CheA, sensor kinase [EC:2.7.13.3] | EFX46098; EFX46097 |
| cheW | CheW, purine-binding protein | EFX46097 | |
| cheX | CheX | Not found | |
| cheC | CheC | EFX44110; EFX46096 | |
| cheD | CheD [EC:3.5.1.44] | EFX46095 | |
| cheR | CheR, methyltransferase [EC:2.1.1.80] | EFX46532 | |
| cheB | CheB, response regulator [EC:3.1.1.61] | EFX46099 | |
| cheY | CheY, response regulator | EFX46498; EFX46108 | |
| cheZ | CheZ | EFX46498 | |
| cheV | CheV, response regulator | EFX46097; EFX46099 | |
| Methyl-accepting chemotaxis proteins | mcp | methyl-accepting chemotaxis protein | EFX44725; EFX44051; EFX45552; EFX46097 |
| tsr | McpI, serine sensor receptor | EFX44051; EFX44725 | |
| tar | McpII, aspartate sensor receptor | EFX44725; EFX46877 | |
| trg | McpIII, ribose and galactose sensor receptor | EFX44725 | |
| tap | McpIV, peptide sensor receptor | Not found | |
| aer | Aerotaxis receptor | EFX44725; EFX46877 | |
| hemAT | haem-based aerotactic transducer | EFX44725; EFX46877 | |
| Type-III secretion proteins for flagellar assembly | fliH | FliH | EFX46029 |
| fliI | FliI, flagellum-specific ATP synthase [EC:3.6.3.14] | EFX46028 | |
| fliZ, fliO | FliO/FliZ | EFX46107 | |
| fliP | FliP, flagellar biosynthetic protein | EFX46106 | |
| fliQ | FliQ, flagellar biosynthetic protein | EFX46105 | |
| fliR | FliR, flagellar biosynthetic protein | EFX46104 | |
| flhA | FlhA, flagellar biosynthesis protein | EFX46102 | |
| flhB | FlhB, flagellar biosynthetic protein | EFX46103 | |
| flhE | FlhE | Not found | |
| flhF | FlhF, flagellar biosynthetic protein | EFX46101 | |
| Motor/Switch | motA | MotA, chemotaxis protein | EFX45489 |
| motB | MotB, chemotaxis protein | EFX45490 | |
| C-ring | fliG | FliG, flagellar motor switch protein | EFX46030 |
| fliM | FliM, flagellar motor switch protein | EFX46110 | |
| fliNY, fliN | FliN/FliY, flagellar motor switch protein | EFX46109 | |
| M, S, P and L rings | fliF | FliF, flagellar M-ring protein | EFX46031 |
| flgI | FlgI, flagellar P-ring protein precursor | not found | |
| flgA | FlgA, flagella basal body P-ring formation protein | not found | |
| flgH | FlgH, flagellar L-ring protein precursor | not found | |
| fliF | FliF, flagellar M-ring protein | EFX46031 | |
| flgA | FlgA, flagella basal body P-ring formation protein | not found | |
| flgH | FlgH, flagellar L-ring protein precursor | not found | |
| Rod, hook, and filament | flgB | FlgB, flagellar basal-body rod protein | EFX46034 |
| flgC | FlgC, flagellar basal-body rod protein | EFX46033 | |
| flgD | FlgD, flagellar basal-body rod modification protein | not found | |
| flgF | FlgF, flagellar basal-body rod protein | EFX46113; EFX45368 | |
| flgG | FlgG, flagellar basal-body rod protein | EFX46113; EFX45367 | |
| flgJ | FlgJ, flagellar protein | not found | |
| flgE | FlgE, flagellar hook protein | EFX46113 | |
| fliE | FliE, flagellar hook-basal body complex protein | EFX46032 | |
| flgK | FlgK, flagellar hook-associated protein 1 | EFX46010 | |
| flgL | FlgL, flagellar hook-associated protein 3 | EFX46011 | |
| fliD | FliD, flagellar hook-associated protein 2 | EFX46016 | |
| fliK | FliK, flagellar hook-length control protein | not found | |
| fliL | FliL, flagellar protein | EFX46111 | |
| fliC | Flagellin | EFX46015 | |
| flaF | FlaF, flagellar protein | EFX46015 | |
| flaG | FlaG, flagellar protein | not found | |
| flbA | FlbA, flagellar protein | EFX46102 | |
| flbB | FlbB, flagellar protein | not found | |
| flbC | FlbC, flagellar protein | not found | |
| flbD | FlbD, flagellar protein | not found | |
| flbT | FlbT, flagellar protein | not found | |
| Regulation | flhC | FlhC, flagellar transcriptional activator | not found |
| flhD | FlhD, flagellar transcriptional activator | not found | |
| fliA | FliA, RNA polymerase sigma factor for flagellar operon | EFX44501;EFX46093; EFX43745; EFX44502 | |
| flgM | FlgM, negative regulator of flagellin synthesis | not found | |
| Chaperones | flgN | FlgN, flagella synthesis protein | not found |
| fliJ | FliJ, flagellar protein | EFX46027 | |
| fliS | FliS, flagellar protein | EFX46017 | |
| fliT | FliT, flagellar protein | not found | |
| fliW | FliW, flagellar assembly factor | EFX46013 | |
| fliY | FliY, cystine transport system substrate-binding protein | EFX46109 | |
| fliZ | FliZ | EFX46107 | |
| Archaeal flagellar proteins | flaA-A, flaA | FlaA, archaeal flagellin | EFX46015 |
| flaB-A, flaB | FlaB, archaeal flagellin | EFX46015 | |
| flaC-A, flaC | FlaC | EFX46015 | |
| flaD-A, flaD | FlaD | EFX46015 | |
| flaE-A, flaE | FlaE | EFX46113 | |
| flaF-A, flaF | FlaF | EFX46015 | |
| flaG-A, flaG | FlaG | not found | |
| flaH-A, flaH | FlaH | not found | |
| flaI-A, flaI | FlaI | not found | |
| flaJ-A, flaJ | FlaJ | not found | |
| flaK-A, flaK | FlaK, archaeal preflagellin peptidase | not found | |
Figure 3.
Flagellar proteins. Distribution of flagellar proteins (excluding chemotaxis proteins) among flagellated Gram-positive (A) and Gram-negative (B) bacteria. The proteins encoded by the P. larvae genome are boxed in green (the ones that are lacking: in red).
Toxins
Our search for P. larvae genes that can encode toxins returned three matches to sixteen proteins (Table 4). A domain of the Clostridum perfringens epsilon toxin (pfam03318) was observed in EFX46729 and EFX45732, which forms pores on host cells [19] leading to cell death. P. larvae appear to possess classic binary toxins: the first factor is a membrane component that makes the host cell permeable to a second factor - the enzymatic component. Seven proteins matched the Clostridial binary toxin B domain (pfam03495), the membrane component. We also saw that four proteins matched the VIP2 domain (cd00233), which is the enzymatic component of a Bacillus toxin capable of ADP ribosylation on actin. In both the Clostridial and Bacillus toxins, the associated partners of these binary toxin-pairs were not observed.
Table 4.
Putative P. larvae toxins.
| Protein (E-value) | Domain, functional information from CDD |
|---|---|
|
EFX46729 (8.00E-20), EFX45732 (1.00E-18) |
ETX_MTX2 [pfam03318]: Clostridium epsilon toxin ETX/Bacillus mosquitocidal toxin MTX2 |
|
EFX43878 (4.00E-32), EFX45096 (3.00E-22), EFX46664 (1.00E-17), EFX46330 (1.00E-10) |
VIP2 [cd00233]: A member of the vegetative insecticidal protein family. An actin-ADP-ribosylating toxin. A binary toxin. |
| EFX43879 (2.00E-102), | Binary_toxB [pfam03495]: Clostridial binary toxin B/anthrax toxin PA. A binary toxin. |
| EFX45058 (1.00E-83), | |
| EFX45095 (2.00E-48), | |
| EFX44748 (3.00E-37), | |
| EFX46332 (2.00E-29), | |
| EFX44749 (6.00E-19), | |
| EFX46331 (1.00E-16), | |
| EFX44747 (4.00E-6), | |
| EFX44951 (6.00E-5), | |
| EFX46847 (6.00E-5), | |
*the listed match is not the one with the lowest E-value; it is displayed because it is the most informative
Hemolysins
In the mid to late stage of the P. larvae infection cycle, bacteria circulate in bee hemolymph which contains hemocytes. Hemolysins are a class of bacterial toxins with lytic activity against blood cells. Four hemolysin domains were matched by proteins from the P. larvae genome. (Table 5). Three proteins contained regions that were highly similar to the hemolytic domain TylC (COG1253) and concomitantly to other domains such as CBS_pair_CorC_HylC_assoc and DUF21 (data not shown), implying that these are very similar proteins. EFX46836 was matched to the hlyIII domain, an integral membrane protein from Bacillus cereus which forms channels on host cells [20]. EFX45686 appears to be related to hemolysin TlyA (TIGR00478) with an indirect pore-forming role. One protein (EFX45724) showed similarity to DUF37 with unpublished claims of hemolysin activity in Aeromonas hydrophila.
Table 5.
Proteins with domains related to hemolysins or cytolytic activity.
| Protein (E-value) | Domain, functional information from CDD |
|---|---|
|
EFX44255 (8.00E-69), EFX44544 (8.00E-72), EFX46152 (3.00E-69) |
TylC [COG1253]: Hemolysins and related proteins |
| EFX46836 (2.00E-23) | hlyIII [pfam03006]: channel-forming cytolysin |
| EFX45724 (1.00E-22) | DUF37 [cl00506]: found in various bacteria, a member from Aeromonas hydrophila has been found to have hemolytic activity (unpublished). |
| EFX45686 (2E-56*) | tly [TIGR00478]: hemolysin TlyA family protein; at least two members of this protein family have been characterized indirectly as pore-forming hemolysins. |
*the listed match is not the one with the lowest E-value; it is displayed because it is the most informative
Proteases
The occurrence of proteases in larval scales (remains of P. larvae-infected bee larvae) has already been well documented [21]. A more recent study hints on a definite possibility that one or more such proteins can act as the virulence factor [22]. Since it has been shown that vegetative rods residing in the host midgut can traverse the epithelium by a paracellular route, P. larvae may generate proteolytic activity that can disrupt epithelial cell-to-cell junctions. We searched the proteins for domains with this function, and after excluding ones with roles that are not directly implicated in pathogenesis (e.g. signal peptidase, SOS response for DNA repair), we list the matches in Table 6. Based on the information provided by the CDD, several proteins may conceivably serve as virulence factors. The Clp protease complex, in particular, has been linked to virulence factor production in Gram-positive bacteria, as well as degradation of misfolded proteins [23,24]. Proteins EFX45851, EFX44792 may be capable of cleaving the transmembrane regions of substrate proteins (matching domains cd06161, cd06158). Given that tight junctions between epithelial cells are mainly composed of occludin and claudin proteins, both of which have transmembrane domains [25], they may be specific substrates for P. larvae proteases. These points, however, are merely speculative, and the many proteases in Table 6 await experimental evidence to prove whether or not they actually contribute a real role in virulence.
Table 6.
Putative P. larvae based on matches to conserved domains.
| Protein (E-value) | Domain, functional information from CDD |
|---|---|
| EFX44344 (2E-37) | AmpS [COG2309]: Leucyl aminopeptidase (aminopeptidase T) |
| EFX45451 (6E-13) | APP-like [cd01092]: Prolidase and Aminopeptidase P |
| EFX47002 (0) | clpC [CHL00095]: Clp protease ATP binding subunit |
| EFX46058 (4E-5) | ClpP [COG0740]: Protease subunit of ATP-dependent Clp proteases |
|
EFX44161 (1E-85), EFX45289 (1E-82) |
clpP [PRK00277]: Clp protease proteolytic subunit |
| EFX45288 (7E-170) | clpX [PRK05342]: Clp protease ATP-binding subunit |
| EFX45551 (7E-82), | COG0826 [COG0826]: Collagenase and related proteases |
| EFX45550 (4E-48), | |
| EFX45481 (3E-19), | |
| EFX44736 (1E-28) | |
| EFX44399 (9E-8*) | COG1310 [COG1310]: Predicted metal-dependent protease of the PAD1/JAB1 superfamily |
| EFX46420 (8E-149) | COG2317 [COG2317]: Zn-dependent carboxypeptidase |
| EFX44401 (9E-45) | COG2738 [COG2738]: Predicted Zn-dependent protease |
|
EFX44533 (2E-9), EFX45796 (2E-9) |
COG2856 [COG2856]: Predicted Zn peptidase |
| EFX46883 (2E-31) | COG3740 [COG3740]: Phage head maturation protease |
| EFX45850 (6E-11) | COG4942 [COG4942]: Membrane-bound metallopeptidase |
|
EFX46250 (1E-34), EFX44926 (2E-64) |
degP_htrA_DO [TIGR02037]: periplasmic serine protease, Do/DeqQ family |
| EFX47184 (5E-45) | DegQ [COG0265]: Trypsin-like serine proteases, typically periplasmic |
| EFX46573 (5E-91), | FtsH_fam [TIGR01241]: ATP-dependent metalloprotease FtsH |
| EFX44535 (5E-20*), | |
| EFX44172 (3E-17*) | |
| EFX47022 (0) | HflB [COG0465]: ATP-dependent Zn proteases |
|
EFX45319 (1E-36*), EFX44073 (5E-8*) |
HflC [COG0330]: membrane protease subunits, stomatin/prohibitin homologs |
| EFX46036 (4E-157) | HslU [COG1220]: ATP-dependent protease HslVU (ClpYQ), ATPase subunit |
| EFX46037 (8E-65) | HslV [COG5405]: ATP-dependent protease HslVU (ClpYQ), peptidase subunit |
| EFX43995 (2E-97), | LasB [COG3227]: Zinc metalloprotease (elastase) |
| EFX45639 (1E-78), | |
| EFX45177 (9E-58) | |
| EFX45284 (0) | Lon [COG0466]: ATP-dependent Lon protease |
| EFX43687 (8E-131) | M3_fam_3 [TIGR02290]: oligoendopeptidase, pepF/M3 family |
| EFX44444 (2E-74) | M14_Endopeptidase_I [cd06229]: Peptidase M14-like domain of Gamma-D-glutamyl-L-diamino acid endopeptidase 1 |
| EFX44181 (2E-45) | MrcA [COG5009]: Membrane carboxypeptidase/penicillin-binding protein |
| EFX45553 (4E-10*), | MrcB [COG0744]: Membrane carboxypeptidase (penicillin-binding protein) |
| EFX46407 (3E-97*), | |
| EFX44298 (3E-75*), | |
| EFX46175 (2E-122*), | |
| EFX44515 (1E-12*), | |
| EFX44516 (4E-11*) | |
| EFX45371 (7E-9), EFX45028 (2E-7), EFX45493 (4E-25) | NlpD [COG0739]: Membrane proteins related to metalloendopeptidases |
| EFX44427 (8E-144) | pepF [TIGR00181]: oligoendopeptidase F |
| EFX46173 (4E-9) | PepN [COG0308]: Aminopeptidase N |
| EFX45105 (1E-66) | PepP [COG0006]: Xaa-Pro aminopeptidase |
| EFX47190 (3E-18) | Peptidase_M23 [pfam01551]: Peptidase family M23 |
| EFX46425 (8E-126) | Peptidase_M6 [pfam05547]: Immune inhibitor A peptidase M6, a metallopeptidase from Bacillus thuringiensis that cleaves host antibacterial proteins |
| EFX47085 (4E-29) | Peptidase_M9_N [pfam08453]: Peptidase family M9 N-terminal |
| EFX46343 (8E-6*) | Peptidase_S24 [pfam00717]: Peptidase S24-like |
| EFX43847 (9E-13) | Peptidase_S41_CPP [cd07560]: C-terminal processing peptidase; serine protease family S41 |
| EFX43943 (3E-27*) | Peptidase_S66 [pfam02016]: LD-carboxypeptidase; this cleaves the bond between an L- and a D-amino acid |
| EFX45480 (6E-38), EFX45479 (2E-8) | Peptidase_U32 [pfam01136]: Peptidase family U32 |
|
EFX43960 (8E-78), EFX45983 (2E-59) |
Peptidases_S8_11 [cd04843]: Peptidase S8 family domain, uncharacterized subfamily 11 |
| EFX46806 (8E-36) | Peptidases_S8_Lantibiotic_specific_protease [cd07482]: Peptidase S8 family domain in Lantiobiotic (lanthionine-containing antibiotics) specific proteases |
| EFX44534 (5E-26), EFX45795 (1E-20*), EFX44174 (8E-25) | Peptidases_S8_Subtilisin_like_2 [cd04847]: Peptidase S8 family domain in Subtilisin-like proteins |
| EFX45938 (8E-46), EFX45020 (3E-62), EFX46180 (2E-50), EFX45070 (2E-20) | Peptidases_S8_Subtilisin_subset [cd07477]: Peptidase S8 family domain in Subtilisin proteins |
|
EFX44839 (5E-67), EFX44567 (5E-47), EFX44567 (4E-55) |
Peptidases_S8_Thermitase_like [cd07484]: Peptidase S8 family domain in Thermitase-like proteins |
| EFX47149 (7E-172) | peptidase-T [TIGR01882]: peptidase T |
| EFX45664 (5E-87) | PepT-like [TIGR01883]: peptidase T-like protein |
|
EFX46054 (6E-33), EFX46055 (2E-7), EFX46066 (2E-67) |
PqqL [COG0612]: Predicted Zn-dependent peptidases |
| EFX45492 (3E-73) | prc [TIGR00225]: C-terminal peptidase (prc) |
| EFX44827 (3E-16*) | PRK00016 [PRK00016]: putative metalloprotease |
| EFX44779 (1E-110) | PRK02858 [PRK02858]: germination protease |
| EFX45078 (5E-118) | PRK07318 [PRK07318]: dipeptidase PepV |
| EFX44480 (4E-11*) | PRK08554 [PRK08554]: peptidase |
| EFX45711 (2E-73) | PRK09795 [PRK09795]: aminopeptidase |
|
EFX45631 (4E-24*), EFX44757 (1E-22*) |
PRK13914 [PRK13914]: invasion associated secreted endopeptidase |
| EFX44490 (3E-22*) | PRK14791 [PRK14791]: lipoprotein signal peptidase |
| EFX46689 (1E-42) | S14_ClpP_1 [cd07016]: Caseinolytic protease (ClpP) |
| EFX44792 (2E-28) | S2P-M50_like_1 [cd06158]: Uncharacterized homologs of Site-2 protease (S2P), zinc metalloproteases (MEROPS family M50) which cleave transmembrane domains of substrate proteins, regulating intramembrane proteolysis (RIP) of diverse signal transduction mechanisms |
| EFX44503 (2E-42) | spore_II_GA [TIGR02854]: sigma-E processing peptidase SpoIIGA |
| EFX45851 (1E-12) | S2P-M50_SpoIVFB [cd06161]: SpoIVFB Site-2 protease (S2P), a zinc metalloprotease (MEROPS family M50B), regulates intramembrane proteolysis (RIP) |
| EFX47001 (2E-8*) | spore_lon_C[TIGR02903]: ATP-dependent protease, Lon family |
| EFX45285 (0) | spore_lonB [TIGR02902]: ATP-dependent protease LonB |
| EFX46081 (7E-45) | TIGR00054 [TIGR00054]: RIP metalloprotease RseP |
| EFX44693 (1E-61) | trio_M42_hydro [TIGR03106]: hydrolase, peptidase M42 family |
*the listed match is not the one with the lowest E-value; it is displayed because it is the most informative
Antibiotic resistance
P. larvae infections have been treated with tetracycline for decades. It has been shown that resistance against this antibiotic can be conferred by the pMA67 plasmid [4]. More recently, the macrolide tylosin has now been registered as a new antibiotic in the U.S. for use when tetracycline is no longer effective [26]. Since antibiotic resistance is inevitable [27], new therapeutics will eventually be required. Realizing that P. larvae appears to encode a plethora of drug efflux proteins (Table 7) may help in the selection of useful drugs.
Table 7.
Putative P. larvae antibiotic resistance proteins based on matches to conserved domains
| Protein (E-value) | Domain, functional information from CDD |
|---|---|
|
EFX43850 (3E-37), EFX47127 (3E-26), EFX43889 (2E-44), |
ABC_BcrA_bacitracin_resist [cd03268]: The BcrA subfamily represents ABC transporters involved in peptide antibiotic resistance; bacitracin is an antibiotic produced by B. licheniformis and B. subtilis with potent antibiotic activity against gram-positive bacteria |
| EFX46602 (1E-57), | |
| EFX44842 (2E-50), | |
| EFX46272 (4E-41), | |
| EFX45233 (9E-52) | |
| EFX46614 (4E-25), | ABC_DR_subfamily_A [cd03230]: This family of ATP-binding proteins belongs to a multisubunit transporter involved in drug resistance (BcrA and DrrA), nodulation, lipid transport, and lantibiotic immunity |
| EFX44699 (4E-16), | |
| EFX45065 (1E-50) | ABC_DrrA [cd03265]: DrrA is the ATP-binding protein component of a bacterial exporter complex that confers resistance to the antibiotics daunorubicin and doxorubicin |
| EFX46465 (1E-55), | ABC_drug_resistance_like [cd03264]: ABC-type multidrug transport system, ATPase component |
| EFX45467 (3E-61), | |
| EFX45475 (2E-48*) | |
| EFX43892 (1E-42), | ABC_putative_ATPase [cd03269]: This subfamily is involved in drug resistance, nodulation, lipid transport, and bacteriocin and lantibiotic immunity |
| EFX46872 (1E-35), | |
| EFX47099 (4E-20*), | |
| EFX45045 (1E-71*) | |
| EFX46236 (3E-6*) | ABCC_MRP_Like [cd03228]: The MRP (Mutidrug Resistance Protein)-like transporters are involved in drug, peptide, and lipid export; they belong to the subfamily C of the ATP-binding cassette (ABC) superfamily of transport proteins. |
| EFX44443 (3E-26) | ACR_tran [pfam00873]: AcrB/AcrD/AcrF family. Members of this family are integral membrane proteins. Some are involved in drug resistance |
| EFX44702 (4E-33), | CcmA [COG1131]: ABC-type multidrug transport system, ATPase component |
| EFX46006 (5E-34), | |
| EFX44595 (4E-16), | |
| EFX43880 (8E-54), | |
| EFX47052 (7E-40), | |
| PL1_2983 (1E-33), | |
| EFX46146 (6E-40), | |
| EFX45253 (3E-22), | |
| EFX45810 (9E-18), | |
| EFX45945 (1E-18), | |
| EFX47082 (9E-21*), | |
| EFX44360 (3E-15*), | |
| EFX46805 (6E-30*), | |
| EFX45180 (5E-27*) | |
| EFX43881 (3E-4*) | COG0842 [COG0842]: ABC-type multidrug transport system, permease component |
| EFX44713 (1E-52) | COG2409 [COG2409]: Predicted drug exporters of the RND superfamily |
| EFX47096 (4E-18) | COG4152 [COG4152]: ABC-type uncharacterized transport system, ATPase component |
| EFX46793 (6E-34) | COG4586 [COG4586]: ABC-type uncharacterized transport system, ATPase component |
| EFX46612 (2E-58), | drrA [TIGR01188]: daunorubicin resistance ABC transporter ATP-binding subunit |
| EFX43786 (8E-63), | |
| EFX45226 (4E-54), | |
| EFX44634 (3E-36*) | |
| EFX43785 (4E-5) | drrB [TIGR01247]: daunorubicin resistance ABC transporter membrane protein; the protein associated with effux of the drug, daunorubicin |
|
EFX44398 (8E-11), EFX47139 (1E-59) |
efflux_Bcr_CflA [TIGR00710]: drug resistance transporter, Bcr/CflA subfamily |
| EFX46169 (4E-40), | efflux_EmrB [TIGR00711]: drug resistance transporter, EmrB/QacA subfamily |
| EFX44387 (7E-37), | |
| EFX44446 (8E-09), | |
| EFX46307(5E-24), | |
| EFX46126 (1E-53), | |
| EFX44956 (3E-9*) | |
| EFX46708 (3E-14) | emrE [PRK09541]: multidrug efflux protein |
| EFX44177 (2E-4*) | Glyoxalase [pfam00903]: Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily |
| EFX45543 (4E-10), | HTH_MARR [smart00347]: helix_turn_helix multiple antibiotic resistance protein |
| EFX43861 (8E-11), | |
| EFX46148 (6E-10), | |
| EFX46170 (4E-12), | |
| EFX44712 (3E-12), | |
| EFX45783 (3E-6*), | |
| EFX46306 (1E-8*), | |
| EFX46227 (3E-4*), | |
| EFX44716 (5E-9*) | |
| EFX45439 (2E-48) | Lant_dehyd_C [pfam04738]: Lantibiotic dehydratase, C terminus |
| EFX45438 (3E-31) | LanC [cd04793]: LanC is the cyclase enzyme of the lanthionine synthetase. Lanthinoine is a lantibiotic, a unique class of peptide antibiotics |
|
EFX46802 (4E-77*), EFX46803 (4E-12*) |
LcnDR2 [COG4403]: Lantibiotic modifying enzyme |
| EFX45784 (1E-84), | MdlB [COG1132]: ABC-type multidrug transport system; ATPase and permease components |
| EFX45333 (6E-107), | |
| EFX44415 (7E-47*), | |
| EFX44416 (5E-44*), | |
| EFX45434 (1E-26*) | |
| EFX45348 (3E-36) | NorM [COG0534]: Na+-driven multidrug efflux pump |
| EFX45532 (1E-114) | PRK01766 [PRK01766]: multidrug efflux protein |
| EFX46249 (6E-16) | PRK03545 [PRK03545]: sugar efflux transporter |
|
EFX46262 (2E-15), EFX44433 (8E-82) |
PRK09874 [PRK09874]: drug efflux system protein MdtG |
| EFX45329 (5E-21) | PRK10054 [PRK10054]: putative MFS-type transporter YdeE |
| EFX47111 (5E-4*) | PRK10504 [PRK10504]: multidrug efflux system protein MdtE |
| EFX45115 (3E-58*) | PRK10522 [PRK10522]: multidrug transporter membrane component/ATP-binding component |
| EFX47080 (1E-129) | PRK10789 [PRK10789]: putative multidrug transporter membrane\ATP-binding components |
| EFX47079 (5E-98) | PRK10790 [PRK10790]: putative multidrug transporter membrane\ATP-binding components |
| EFX45606 (2E-13) | PRK11102 [PRK11102]: bicyclomycin/multidrug efflux system |
|
EFX46841 (4E-19), EFX46842 (3E-13) |
PRK11431 [PRK11431]: quaternary ammonium compound-resistance protein SugE |
| EFX43805 (8E-7*) | PRK11646 [PRK11646]: multidrug resistance protein MdtH |
| EFX47100 (4E-6) | RND_mfp [TIGR01730]: RND family efflux transporter, MFP subunit |
*the listed match is not the one with the lowest E-value; it is displayed because it is the most informative
It is known that the bee midgut contains a vast array of bacteria, some of which can inhibit P. larvae growth in vitro [28-30], presumably by the production of lantibiotics. In our search we were able to find a number of different drug and lantibiotic efflux pumps and modifying enzymes (Table 7), and it is likely that these proteins are enlisted by P. larvae during interactions with co-occurring antagonistic bacteria.
Conclusions
In this article we report an update on the P. larvae genome to 182-fold coverage, and estimated the ordering of contigs based on comparison against the P. JDR2 genome. We predicted more than 3500 genes and provided these with functional annotation. Of great interest are enzymes that allow the bacterium to traverse the midgut epithelium after ingestion, a process which can contribute to its virulence. Proteases have been thought of as a key factor [22], and this is supported by the large repertoire of such enzymes which we have seen from the predicted gene products. Damage to the host is likely the result of mixed effects from the hemolysins and toxins that can be encoded by P. larvae. Its ability to survive in the host by evading the immune system, as well as protecting itself against other gut bacteria, is likely made possible due to the antibiotic proteins that may be expressed.
This improved version of the P. larvae genome and the more detailed annotations will be tremendously useful for improving our understanding of this species; for example, in designing primers to clone genes, or mutate them by site-directed mutagenesis. Especially for the field of proteomics, which often relies heavily on a database of protein sequences to make identifications, the current genome update will be a very powerful tool. Computer-based modeling techniques can be employed to predict the structure of virulence factors, which can guide drug design. Ultimately, the genome will pave the way to more effective prevention; or better yet, a cure for AFB. Given the bee population is under severe strain from the widely publicized Colony Collapse Disorder, attributable to multiple biotic and abiotic factors, it is important to continually improve our knowledge of the key threats that burden worldwide bee health.
Methods
Preparation of genomic DNA
P. larvae strain BRL-230010, stored at -80°C was cultured for 5 d in a shaking incubator at 35°C in 25 mL of media. This media contained 1% Yeast Extract (Difco), 1% soluble starch which was prepared in 10 mM KH2PO4 and adjusted to pH 6.6 with KOH (Recipe #4 from [31]). Genomic DNA was extracted using the DNeasy kit (Qiagen), following the manufacturer's protocol for Gram-positive bacteria.
Genome sequencing
8,212,402 2 × 50 bp paired-end reads with a mean fragment size of 182 bp were sequenced at the Genome Sciences Centre, Vancouver, Canada using the Illumina GAIIx platform as per manufacturer's instructions. This yielded approximately 182-fold coverage of the estimated genome size. In addition, 24,768 Sanger capillary-derived sequences were generated at the Human Genome Sequencing Centre, Baylor College of Medicine, using the ABI3730 sequencing platform. This provided an additional 3-fold coverage. Raw Sanger sequences were aligned against the UniVec database (NCBI http://www.ncbi.nlm.nih.gov) using BLAST. Those Sanger sequences that had good BLAST hits (e-value < 1e-10) were filtered from the dataset. Then, vector sequences were trimmed from the raw Sanger reads using cross_match (Cross_match http://www.phrap.org/) against the UniVec database. We then aligned all the Illumina reads to the Sanger sequences using MAQ version 0.7.1 [32]. Due to observed Honey Bee DNA contamination arising within the Sanger sequences, only sequences where at least 30% of the length aligned with Illumina reads were retained.
In the first stage of the assembly process, the Illumina reads were assembled into contigs using ABySS version 1.2.7 [33]. Assemblies were attempted using several different values for the ABySS sequence overlap parameter k (which determines the k-mer length used to construct the assembly graph), with the best assembly found at k = 37. Other modified ABySS parameter values were s = 100, n = 10, and d = 50. All other ABySS parameters were set to their default values. Following the initial assembly, contigs were joined into scaffolds by aligning the long-insert Sanger sequence data to the assembly, and merged using the scaffolding module of ABySS (Jackman et al., in preparation). Where possible, scaffold gaps were filled, and contigs were extended, using the local reassembly tool Anchor (version 0.2.7, http://www.bcgsc.ca/platform/bioinfo/software/anchor/releases/0.2.7) (Docking et al., in preparation).
Genome annotation
ORFs were predicted with GLIMMER [34] using the long-ORF training script provided with the software package. ORFs were annotated as predicted proteins if they have at least 100 amino acids or at least 50 with a BLAST hit at a threshold of E = 10-5 to Bacillus or Streptococcus (Additional File 1). Annotation terms were transferred from FASTA headers where appropriate, and in accordance with NCBI Prokaryotic Genome Submission Guidelines (full list of proteins in Additional File 2).
Genomic dot-plot comparisons were made with the program MUMmer [35]. To produce Figure 2, P. larvae contigs were ordered manually based on BLAST hit coordinates to P. JDR2 and P. vortex as well as the graphical output of MUMmer alignments.
The genome annotation as described above was accomplished using the first version of the assembly from this sequencing (ADZY01000000), which had a total N50 of 56,120 base pairs, a total length of 4,352,422, and a mean contig size of 12,329 bp. When we repeated this analysis with the latest assembly, all but 53 of the 3,568 gene models were identical. Of the 53, most were strong partial hits. All downstream analysis was done using ADZY01000000.
Shotgun proteomics of P. larvae lysates
P. larvae culture was prepared as above. Bacteria were collected by centrifugation for 5 minutes at 16100 relative centrifugal force and the pellet was washed twice in PBS. For an in-gel digestion, the bacteria were lysed in 100 μl of 4× LDS and heated for 15 minutes at 90°C, passed 5 times through a 26 G needle fitted with a syringe before heating for another 10 minutes at 90°C. Lysis was observed under the microscope. Protein concentration was estimated by Coomassie Plus staining, and 200 μg of protein was divided among all 10 wells of a 2-12% NuPAGE precast MES mini gel set up according to the manufacturer's instructions and the in-gel digestion proceeded essentially as described in [36]. For in-solution digestions, bacteria was lysed by either 100 μl of 6 M urea, 2 M thiourea, 20 mM Tris-HCl, pH 8.0 or 0.2 μl RapiGest in 100 μl NH4HCO3 pH 8 and boiled for 6 minutes. Bacteria from both samples were lysed by the needle and syringe method as above. In a third sample, bacterial surface proteins were stripped by adding 100 μL of 100 mM glycine pH 2 and proteins precipitated by ethanol before being denatured using urea/thiourea as above. Proteins were reduced, alkylated, digested with LysC (urea sample only) and trypsin and purified as described in [36]. Proteins from all three samples, as well as gel fragments, were fractionated by strong cation exchange STAGE tips [37]. Peptides were subjected to liquid chromatography-tandem mass spectrometry on an LTQ-Orbitrap and FT-ICR (Thermo) according to procedures described in [36]. The raw data were searched against the new P. larvae protein set derived as above using MaxQuant (v1.1.1.25) [38], considering up to two missed cleavages, Cys carbamidomethylation as a fixed modification and Met oxidation and N-terminal acetylation as variable modifications. The overall mass accuracy of the data set was 0.99 ppm and the false discovery rate, at the protein level, was set to 1%.
Bioinformatic annotation of predicted proteins
Predicted proteins were BLAST-searched against selected amino acid sequences using the stand-alone BLAST-software package [12]. For the defined targets, i.e. 'flagellar system', 'toxins', 'hemolysins' and 'proteases', query files of representative protein sequences in FASTA format were composed by key word searching the 'protein' database of the National Center of Biotechnology Information (NCBI) at http://www.ncbi.nlm.nih.gov/. The outcome of each BLASTp-search was verified by cross searching the found hits on the online BLASTp tool (NCBI) and by an in depth, manual comparison of their conserved domain architecture and sequence identity. To expand our search for relevant domains, we used BLASTp to search the CDD [39] which contain domain sequences from various sources (Pfam v 24, COG v1, SMART v5.1, Entrez protein clusters database, TIGRFAM v9.0, NCBI, Entrez multi-model superfamilies), with the maximum E-value set at 10-3. Where there are matches to more than one domain, we only highlight the one with the lowest E-value in the manuscript. Full results are shown in Additional File 4.
Authors' contributions
QWTC did all of the wet-bench work, coordinated the whole project and led the writing of the manuscript. SJMJ oversaw the sequencing and assembly. IB, NYL, SKC, SDJ, TRD and GAT conducted all the genome sequencing and assembly. RSC conducted contig ordering. QWTC, RSC, and DCdG did the function annotation and analysis. JDE and LJF had the initial idea for this project and guided the analyses. All authors helped to write the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Annotation of P. larvae proteins. This table provides annotation information for predicted P. larvae proteins found using BLAST searches against Bacillus or Streptococcus.
Predicted gene models for the P. larvae genome. The information here is generated from GLIMMER, which was used on the P. larvae genome to predicted gene models.
Mass spectrometry-sequenced P. larvae peptides. Shown here are shotgun proteomics sequenced peptides of P. larvae lysates, which serve as experimental evidence for predicted proteins.
Functional annotation of predicted P. larvae proteins based on matches to conserved domains. Protein sequences of P. larvae (column A) were searched against the Conserved Domain Database by BLAST. All matches with an Expect Value (column F) of less than 10-3 are reported here (column B, C), with the hit having the lowest Expect Value marked in column G. A brief description of the domains' function is shown in column D and E. Additional qualifiers such as percent identity, match length, score are found in columns H to P.
Contributor Information
Queenie WT Chan, Email: queeniecwt@shaw.ca.
R Scott Cornman, Email: scott.cornman@gmail.com.
Inanc Birol, Email: ibirol@bcgsc.ca.
Nancy Y Liao, Email: nliao@bcgsc.ca.
Simon K Chan, Email: sichan@bcgsc.ca.
T Roderick Docking, Email: rdocking@bcgsc.ca.
Shaun D Jackman, Email: sjackman@bcgsc.ca.
Greg A Taylor, Email: gtaylor@bcgsc.ca.
Steven JM Jones, Email: sjones@bcgsc.ca.
Dirk C de Graaf, Email: Dirk.deGraaf@UGent.be.
Jay D Evans, Email: Jay.Evans@ARS.USDA.GOV.
Leonard J Foster, Email: foster@chibi.ubc.ca.
Acknowledgements
The authors wish to thank the members of their respective groups for helpful guidance and discussions; in particular, Nikolay Stoynov for technical assistance with mass spectrometry. This work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant to LJF. LJF is the Canada Research Chair in Quantitative Proteomics. The infrastructure used in this work was supported, in part, by the Canada Foundation for Innovation, the BC Knowledge Development Fund and the BC Proteomics Network.
References
- Shimanuki H. In: Honey bee pests, predators, and diseases. Morse RA, Flottum K, editor. Medina: A.I. Root Co.; 1997. Bacteria; pp. 35–54. [Google Scholar]
- Brødsgaard CJ, Ritter W, Hansen H. Response of in vitro reared honey bee larvae to various doses of Paenibacillus larvae larvae spores. Apidologie. 1998;29:569–578. doi: 10.1051/apido:19980609. [DOI] [Google Scholar]
- Yue D, Nordhoff M, Wieler LH, Genersch E. Fluorescence in situ hybridization (FISH) analysis of the interactions between honeybee larvae and Paenibacillus larvae, the causative agent of American foulbrood of honeybees (Apis mellifera) Environ Microbiol. 2008;10(6):1612–20. doi: 10.1111/j.1462-2920.2008.01579.x. [DOI] [PubMed] [Google Scholar]
- Murray KD, Aronstein KA, de Leon JH. Analysis of pMA67, a predicted rolling-circle replicating, mobilizable, tetracycline-resistance plasmid from the honey bee pathogen, Paenibacillus larvae. Plasmid. 2007;58(2):89–100. doi: 10.1016/j.plasmid.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Qin X, Evans JD, Aronstein KA, Murray KD, Weinstock GM. Genome sequences of the honey bee pathogens Paenibacillus larvae and Ascosphaera apis. Insect Mol Biol. 2006;15(5):715–8. doi: 10.1111/j.1365-2583.2006.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirota-Madi A, Olender T, Helman Y, Ingham C, Brainis I, Roth D, Hagi E, Brodsky L, Leshkowitz D, Galatenko V, Nikolaev V, Mugasimangalam RC, Bransburg-Zabary S, Gutnick DL, Lancet D, Ben-Jacob E. Genome sequence of the pattern forming Paenibacillus vortex bacterium reveals potential for thriving in complex environments. BMC Genomics. 2010;11:710. doi: 10.1186/1471-2164-11-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikiori T, Naganawa H, Muraoka Y, Aoyagi T, Umezawa H. Plipastatins: new inhibitors of phospholipase A2, produced by Bacillus cereus BMG302-fF67. III. Structural elucidation of plipastatins. J Antibiot (Tokyo) 1986;39(6):755–61. doi: 10.7164/antibiotics.39.755. [DOI] [PubMed] [Google Scholar]
- Arima K, Kakinuma A, Tamura G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochem Biophys Res Commun. 1968;31(3):488–94. doi: 10.1016/0006-291X(68)90503-2. [DOI] [PubMed] [Google Scholar]
- Sen R. Surfactin: biosynthesis, genetics and potential applications. Adv Exp Med Biol. 2010;672:316–23. doi: 10.1007/978-1-4419-5979-9_24. [DOI] [PubMed] [Google Scholar]
- Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18(4):380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- Dale SE, Sebulsky MT, Heinrichs DE. Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J Bacteriol. 2004;186(24):8356–62. doi: 10.1128/JB.186.24.8356-8362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Waltman P, Kacmarczyk T, Bate AR, Kearns DB, Reiss DJ, Eichenberger P, Bonneau R. Multi-species integrative biclustering. Genome Biol. 2010;11(9):R96. doi: 10.1186/gb-2010-11-9-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, Goto S, Kanehisa M. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008. pp. W423–6. [DOI] [PMC free article] [PubMed]
- Liu R, Ochman H. Stepwise formation of the bacterial flagellar system. Proc Natl Acad Sci USA. 2007;104(17):7116–21. doi: 10.1073/pnas.0700266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Ohto Y, Aizawa S, Macnab RM, Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J Bacteriol. 1994;176(8):2272–81. doi: 10.1128/jb.176.8.2272-2281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer R, Chen PY, Armitage JP, Reinert G, Deane CM. Deciphering chemotaxis pathways using cross species comparisons. BMC Syst Biol. 2010;4:3. doi: 10.1186/1752-0509-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Kojima S, Homma M. Flagellar motility in bacteria structure and function of flagellar motor. Int Rev Cell Mol Biol. 2008;270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- Petit L, Maier E, Gibert M, Popoff MR, Benz R. Clostridium perfringens epsilon toxin induces a rapid change of cell membrane permeability to ions and forms channels in artificial lipid bilayers. J Biol Chem. 2001;276(19):15736–40. doi: 10.1074/jbc.M010412200. [DOI] [PubMed] [Google Scholar]
- Baida GE, Kuzmin NP. Cloning and primary structure of a new hemolysin gene from Bacillus cereus. Biochim Biophys Acta. 1995;1264(2):151–4. doi: 10.1016/0167-4781(95)00150-f. [DOI] [PubMed] [Google Scholar]
- Dancer BN, Chantawannakul P. The Proteases of American Foulbrood Scales. J Invertebr Pathol. 1997;70(2):79–87. doi: 10.1006/jipa.1997.4672. [DOI] [PubMed] [Google Scholar]
- Antunez K, Anido M, Evans JD, Zunino P. Secreted and immunogenic proteins produced by the honeybee bacterial pathogen, Paenibacillus larvae. Vet Microbiol. 2009. [DOI] [PubMed]
- Frees D, Qazi SN, Hill PJ, Ingmer H. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol Microbiol. 2003;48(6):1565–78. doi: 10.1046/j.1365-2958.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- Kruger E, Witt E, Ohlmeier S, Hanschke R, Hecker M. The clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J Bacteriol. 2000;182(11):3259–65. doi: 10.1128/JB.182.11.3259-3265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Yamazaki Y, Katsuno T, Tamura A, Tsukita S. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene. 2008;27(55):6930–8. doi: 10.1038/onc.2008.344. [DOI] [PubMed] [Google Scholar]
- Alippi AM, Albo GN, Reynaldi FJ, De Giusti MR. In vitro and in vivo susceptibility of the honeybee bacterial pathogen Paenibacillus larvae subsp. larvae to the antibiotic tylosin. Vet Microbiol. 2005;109(1-2):47–55. doi: 10.1016/j.vetmic.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Woodford N, Livermore DM. Infections caused by Gram-positive bacteria: a review of the global challenge. J Infect. 2009;59(Suppl 1):S4–16. doi: 10.1016/S0163-4453(09)60003-7. [DOI] [PubMed] [Google Scholar]
- Carina Audisio M, Torres MJ, Sabate DC, Ibarguren C, Apella MC. Properties of different lactic acid bacteria isolated from Apis mellifera L. bee-gut. Microbiol Res. 2011;166(1):1–13. doi: 10.1016/j.micres.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Evans JD, Armstrong TN. Antagonistic interactions between honey bee bacterial symbionts and implications for disease. BMC Ecol. 2006;6:4. doi: 10.1186/1472-6785-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren E, Olofsson TC, Vásquez A, Fries I. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie. 2010;41(1):99–108. doi: 10.1051/apido/2009065. [DOI] [Google Scholar]
- Bailey L, Lee DC. Bacillus larvae: its cultivation in vitro and its growth in vivo. J Gen Microbiol. 1962;29:711–717. doi: 10.1099/00221287-29-4-711. [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19(6):1117–23. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–9. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan QW, Howes CG, Foster LJ. Quantitative comparison of caste differences in honeybee hemolymph. Mol Cell Proteomics. 2006;5(12):2252–62. doi: 10.1074/mcp.M600197-MCP200. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Mann M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J Proteome Res. 2006;5(4):988–94. doi: 10.1021/pr050385q. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011. pp. D225–9. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Annotation of P. larvae proteins. This table provides annotation information for predicted P. larvae proteins found using BLAST searches against Bacillus or Streptococcus.
Predicted gene models for the P. larvae genome. The information here is generated from GLIMMER, which was used on the P. larvae genome to predicted gene models.
Mass spectrometry-sequenced P. larvae peptides. Shown here are shotgun proteomics sequenced peptides of P. larvae lysates, which serve as experimental evidence for predicted proteins.
Functional annotation of predicted P. larvae proteins based on matches to conserved domains. Protein sequences of P. larvae (column A) were searched against the Conserved Domain Database by BLAST. All matches with an Expect Value (column F) of less than 10-3 are reported here (column B, C), with the hit having the lowest Expect Value marked in column G. A brief description of the domains' function is shown in column D and E. Additional qualifiers such as percent identity, match length, score are found in columns H to P.