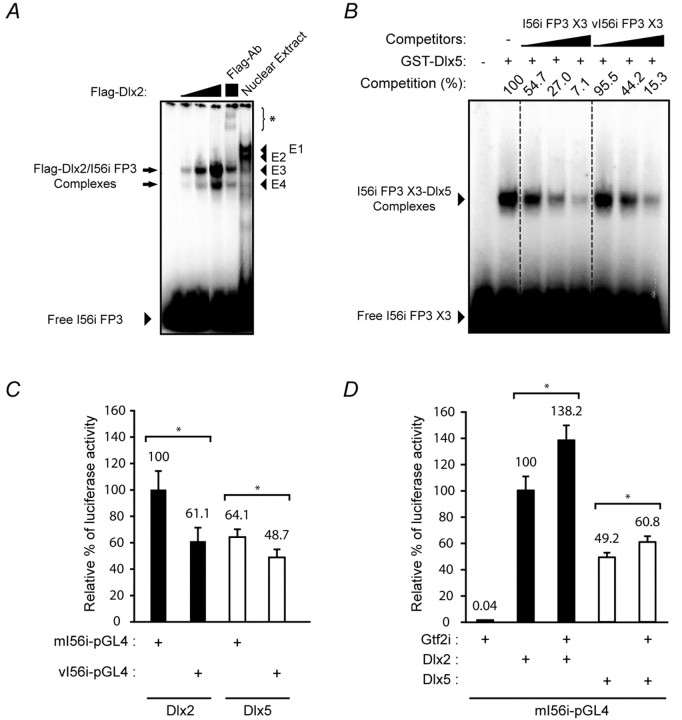
Fig. 6.
Transcriptional activation of I56i by Dlx and Gtf2i is impaired by the 182-SNP. (A) The molecular weight of the protein component(s) of the E4 complex is similar to that of Dlx2. EMSA using the I56i FP3 oligonucleotide with E13.5 mouse embryonic forebrain nuclear extract (Nuclear Extract) or increasing amounts of recombinant Flag-tagged Dlx2 (Flag-Dlx2). Preincubation of the recombinant Dlx2 with an anti-Flag antibody leads to the formation of a high molecular weight complex (asterisk, Flag-Ab lane). (B) Affinity of Dlx proteins for the FP3 site is affected by the 182-SNP. EMSA was performed using the I56i X3 oligonucleotide and a fixed amount of recombinant Dlx5. The resulting complexes (arrowhead) were competed more efficiently by the unlabeled wild-type I56i X3 (increasing amounts in lanes 3 to 5) than by the variant (vI56i X3, lanes 6 to 8) competitor. Competition (%) refers to the percentage of remaining complexes with respect to the control reaction (without competitor, 100%). (C) Replacement of I56i by the 182-SNP-containing variant (vI56i-pGL4.23) impaired transcriptional activation by Dlx2 or Dlx5. (D) Dlx2 and Dlx5 synergize with Gtf2i in activating the transcription of a luc2 reporter construct containing the I56i enhancer. Values shown in C and D represent the mean of relative luciferase activity obtained from five (C) or six (D) independent experiments ± s.e.m. Relative luciferase activity was normalized to Dlx2 plus mI56i-pGL4 (100%). *, P<0.05.