Figure 2.
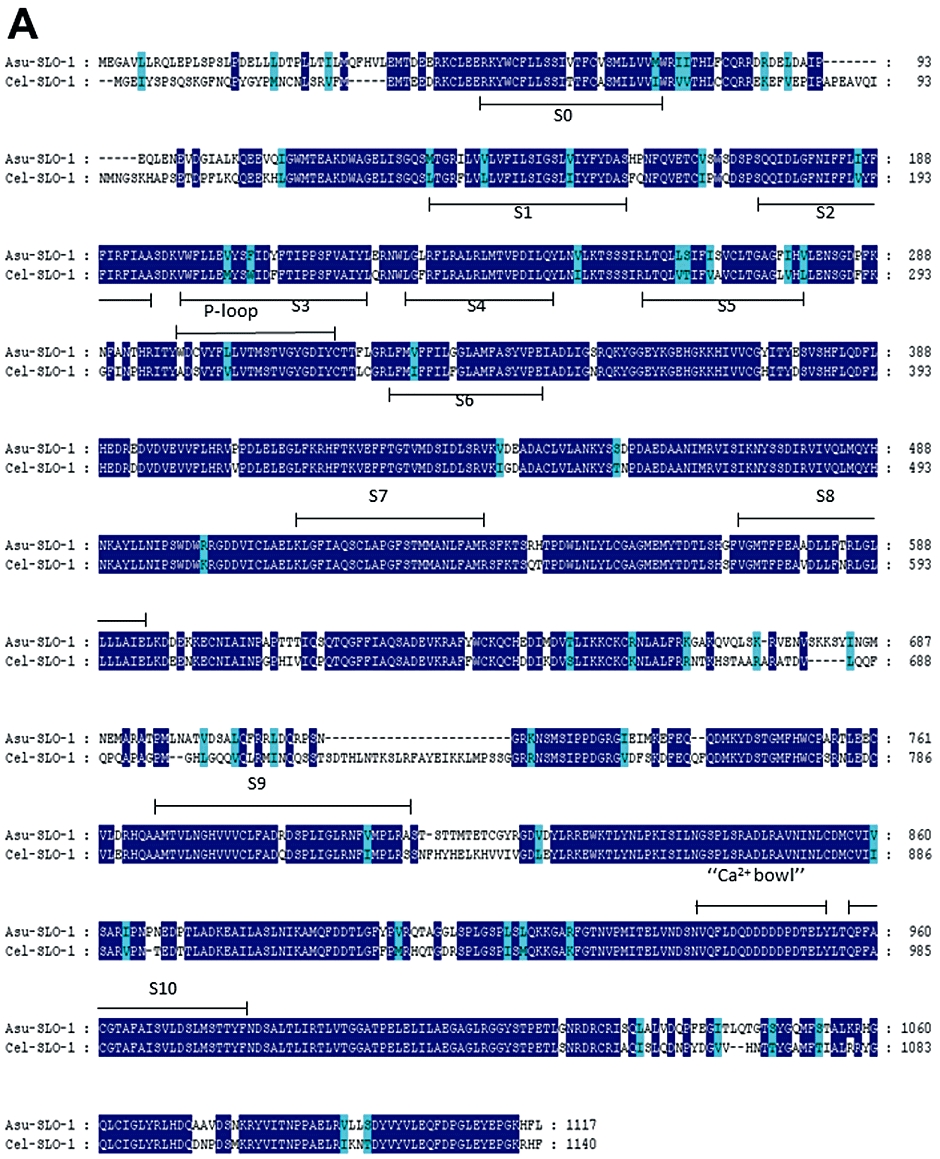
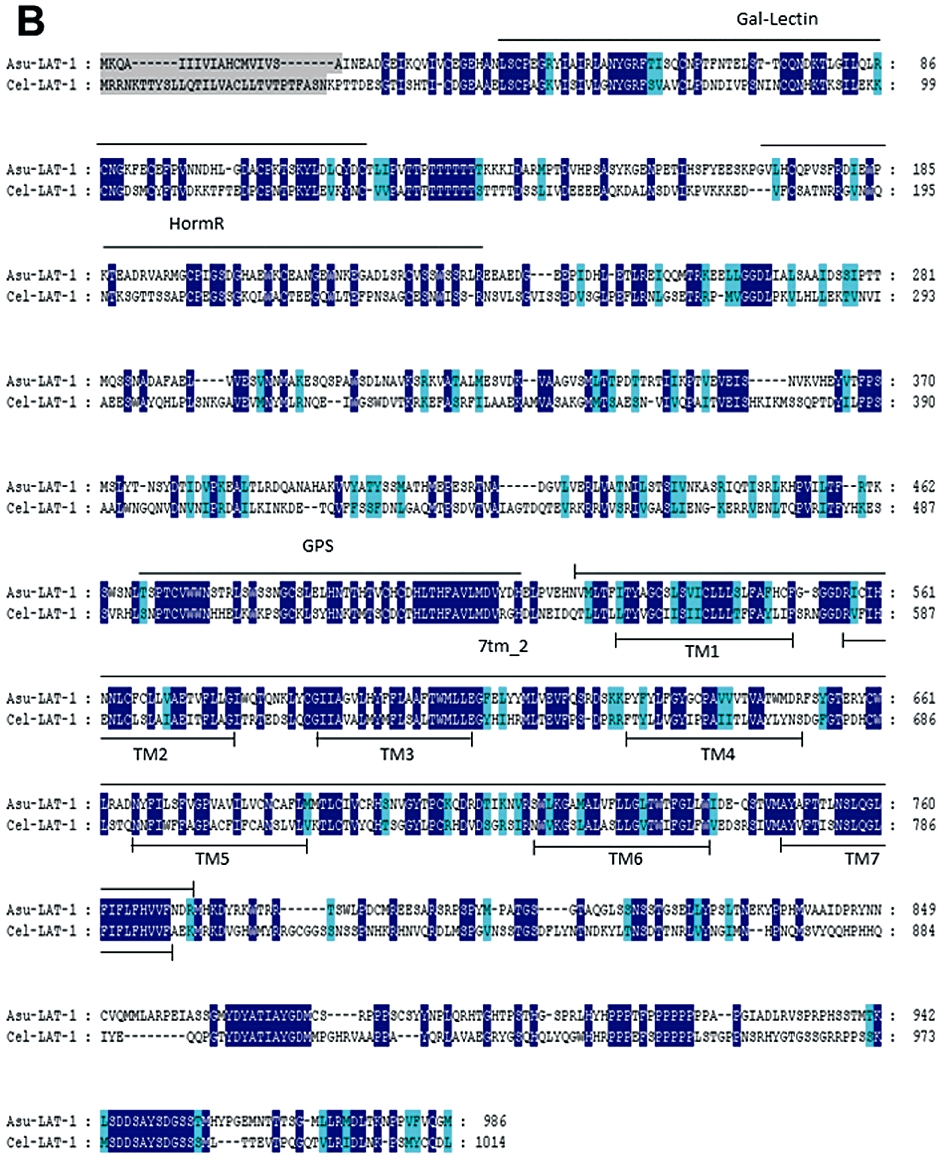
Conservation of SLO-1 and LAT-1 sequences in C. elegans and A. suum. Alignments of deduced amino-acid sequences were constructed using the MUSCLE algorithm (Edgar, 2004) and further processed using GeneDoc program (http://www.nrbsc.org/gfx/genedoc/index.html). Identical amino-acids between C. elegans and A. suum sequences are shaded in dark blue and distinct amino-acids sharing similar physico-chemical properties are shaded in light blue. Predicted signal peptide sequences are shaded in grey. The transmembrane domains (TM) are noted below the sequences. (A) Comparison of Asu-SLO-1 (GenBank accession n° ACC68842.1) and Cel-SLO-1 (GenBank accession n° NP_001024259.1) showing the 7 TM domains (S0–S6), P-loop, S7–S10 intracellular domains and ‘Ca2+ bowl’. Domain annotation corresponds to A. suum SLO-1. (B) Comparison of Asu-LAT-1 (GenBank accession n°ADY40714.1) and Cel-LAT-1 (GenBank accession n°NP_495894.1). Domain annotations correspond to A. suum LAT-1.
