Figure 1.
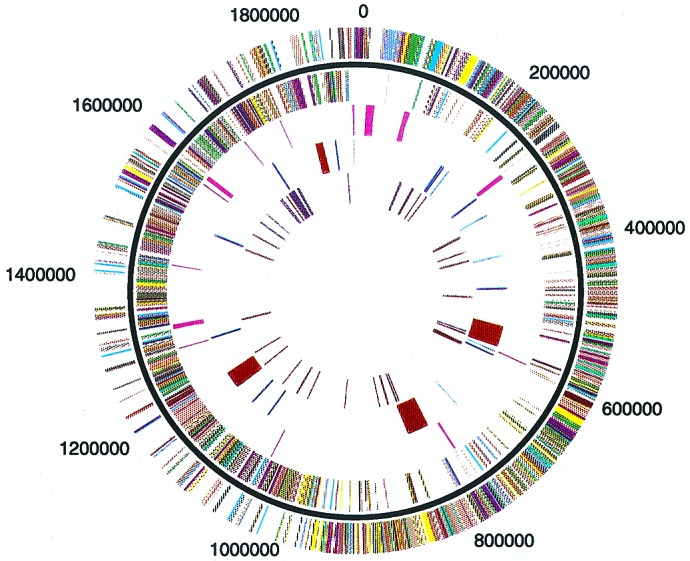
Circular representation of the S. pyogenes strain SF370 genome. Outer circle, predicted coding regions transcribed on the forward (clockwise) DNA strand. Second circle, predicted coding regions transcribed on the reverse (counterclockwise) DNA strand. Third circle, stable RNA molecules. Fourth circle, mobile genetic elements: burgundy, bacteriophage; blue, transposons/IS elements; light cyan, transposons/IS elements (pseudogenes). Fifth circle, known and putative virulence factors: purple, previously identified ORFs; brown, ORFs identified as a result of genome sequence. The lines in each concentric circle indicate the position of the represented feature. Colors: dark gray, amino acid transport and metabolism; light gray, carbohydrate transport and metabolism; green, cell division and chromosome portioning; olive green, cell envelope biogenesis, outer membrane; salmon, cell motility and secretion; tan, coenzyme metabolism; violet, DNA replication, recombination and repair; yellow, energy production and conversion; light pink, function unknown; rose, general function prediction only; light brown, inorganic ion transport and metabolism; light purple, lipid metabolism; light blue, nucleotide transport and metabolism; orange, posttranslational modification, protein turnover, chaperones; red, signal transduction mechanisms; cyan, transcription; green, translation, ribosomal structure and biogenesis; purple, virulence factors; magenta, stable RNA; burgundy, bacteriophage; medium blue, pseudogenes; brown, newly identified virulence factors; blue, transposons/IS elements.
