Figure 3.
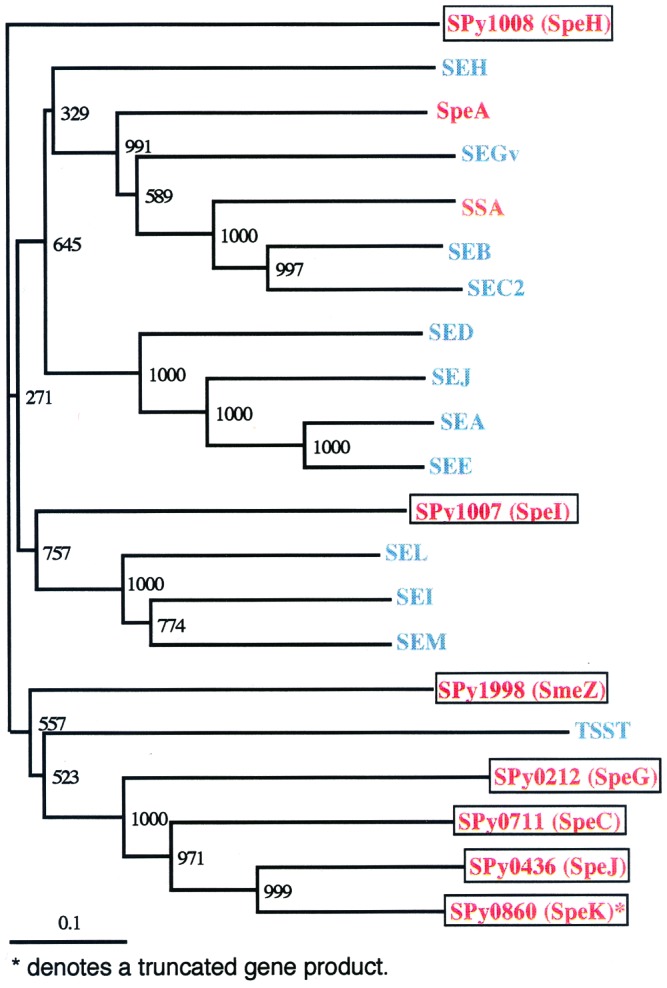
Phylogram of superantigen-like proteins identified in S. pyogenes SF370. The protein alignment was generated by using clustalx (by using the blossum matrix and a bootstrap trial of 1,000). The graphical representation of the tree was generated by using treeview. Gene products encoded by SF370: red; encoded by S. pyogenes but not present in SF370: green and encoded by S. aureus. Scale bar represents the length of the branches. Bootstrap values are displayed at each internal node. Note: SpeK is present in SF370 as a partial product only; an intact copy of speK has not yet been identified in S. pyogenes. Gene products encoded by S. pyogenes are in red with those proteins specifically encoded by strain SF370 also enclosed in a box. The products encoded by S. aureus are in blue. GenBank accession nos.: S. pyogenes proteins: SSA (gb: AAA65928.1); SpeA (gb: AAC48868.1). S. aureus proteins: SEA (prf:1704203A); SEB (gb: AAA88550.1); SEC2 (gb: AAA26624.1); SED (gb: AAB06195.1); SEE (gb: AAA26617.1); SEGv (dbj: BAA36693.1); SEH (gb: AAA19777.1); SEI (gb: AAC26661.1); SEJ (gb: AAC78590.1); SEL (gb: AAG29598.1); SEM (gb: AAG36952.1); TSST (gb: AAA26682.1). Supplementary information is available on the world wide web sites for our laboratories at the University of Oklahoma (http://www.genome.ou.edu/) and the University of Oklahoma Health Sciences Center (http://www.microgen.ouhsc.edu/).
