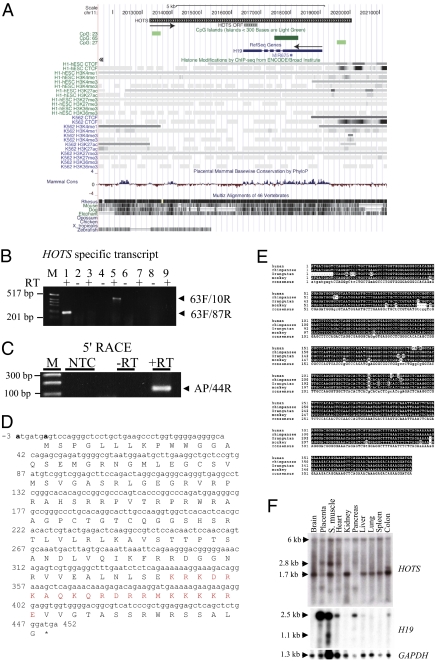
Fig. 1.
HOTS is a primate-conserved, ubiquitous transcript antisense to H19. (A) Genomic organization of HOTS and H19 genes on human chromosome 11p15.5. The direction of HOTS and H19 transcriptional orientation is shown by black arrows. HOTS ORF is shown. The display is populated with CpG island (green) CTCF-binding sites, ENCODE histone modification marks, and mammalian DNA sequence conservation for the region [University of California at Santa Cruz (UCSC) on Human Genome Sequence: Feb. 2009 (GRCh37/hg19) Assembly, http://genome.ucsc.edu/]. (B) RT-PCR amplification of HOTS using strand-specific RT primer 1119R located within the first intron of H19, followed by amplification with intronic PCR primer pairs 63F/87R (lanes 1 and 2) and 63F/10R (lanes 5 and 6). RT-PCR amplification of H19 using strand-specific RT primer 1F, followed by intronic PCR primer pair 63F/87R (lanes 3 and 4) or 63F/10R (lanes 7 and 8). (Lane 9) No RT primer included but RT enzyme and 63F/87R primer pair. (C) 5′ RACE using primer 4534R for RT and primers 44R and PCR anchor primer. M, 1-kb DNA ladder; NTC, no template control. (D) Sequence for the HOTS ORF, nuclear localization signal shown in red type and the putative Kozak consensus sequences at −3 (A) and +4 (A) in boldface type. (E) Multiple sequence alignment of predicted HOTS primates sequences from human, chimpanzee, orangutan, and monkey. Sequence areas depicted against a black background represent identity, and small letters in the consensus sequence at the bottom show areas of sequence variability. (F) HOTS and H19 human multiple tissues Northern blots. (Top) HOTS. (Middle) H19. (Bottom) GAPDH.