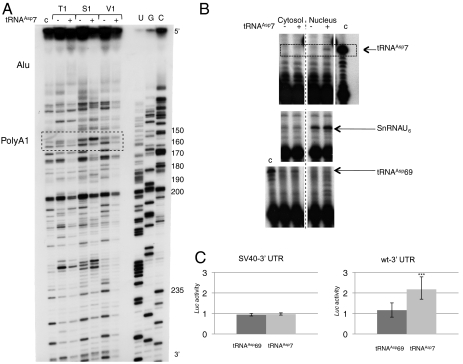
Fig. 5.
Functional study of tRNAAsp7. (A) Long-range rearrangement of AspRS 3′ UTR upon tRNAAsp7 binding. Autoradiogram of 12% denaturing gels displaying probing experiments performed on nt 100 to 250 of the 3′ UTR (in the context of the full-length AspRS mRNA), in the absence (−) or presence (+) of tRNAAsp7. Experiments were performed with RNases T1 (0.4 U), S1 (5.4 × 10-3 U) and V1 (8.3 × 10-3 U). Sequencing (C, G, U) (62) allowed numbering. The dashed box highlights the position of the proximal poly(A) signal. The Alu insertion, which binds tRNAAsp7, is also indicated about 100 nt upstream (top of the autoradiogram). (B) Nuclear localization of tRNAAsp7. RNA molecules were detected by premature chain termination primer extension, which allows elongation of radio-labeled specific primers by 10, 11 and 8 nt matching tRNAAsp69, tRNAAsp7 and SnRNAU6, respectively. Controls (c) were run on in vitro transcribed tRNAAsp7 and tRNAAsp69. Nuclear and cytosolic fractionation is identified by the presence of SnRNAU6 and tRNAAsp69, respectively. (C) Luciferase activity. Hepa 1–6 cells were cotransfected with pRL-CMV (SV40-3′ UTR) or pRL-CMV-WT-3′ UTR (WT-3′ UTR) and pSK-tRNAAsp69 or pSK-tRNAAsp7 and assayed for luciferase activity (deviation values of four independent transfections). Errors bars indicating the standard deviation are given. *** shows statistical significance (tRNAAsp7 versus tRNAAsp69) at P = 0.0006 (Welch’s t test).