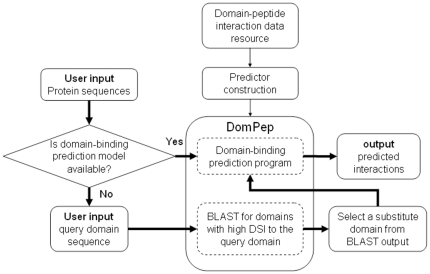
Figure 5. Framework of the DomPep program.
DomPep contains three components- a Graphical user interface (GUI), a local BLAST server and a set of embedded domain-peptide binding predictors. The GUI allows a user to input protein sequences and choose predictors for specific domains. If the query domain does not have an embedded predictor, a substitute query domain with a known predictor can be identified from domains that exhibit the highest DSI from a BLAST search. The output of a DomPep prediction consists of a list of peptides with prediction scores arranged in a descending order from top to bottom. The corresponding proteins are listed in a separate column.