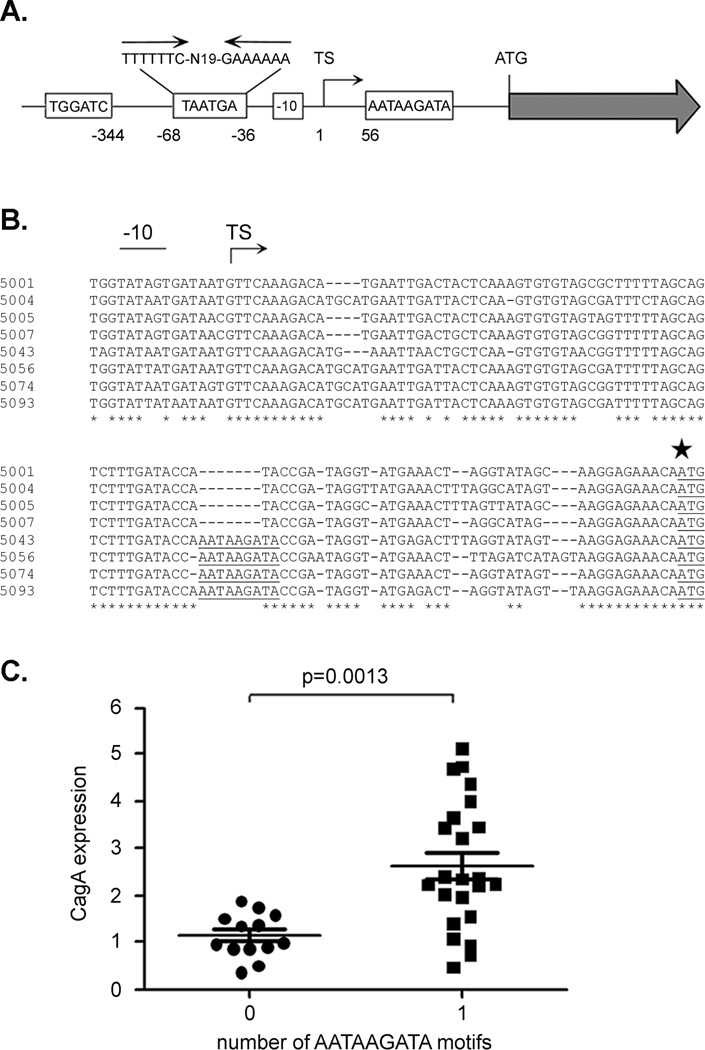
Figure 2. Analysis of an AATAAGATA motif.
A. Regions upstream from cagA that exhibit substantial sequence heterogeneity among Colombian H. pylori strains are illustrated. The Colombian strains vary in the number of copies of TGGATC, TAATGA, and AATAAGATA motifs. The TAATGA motifs are located within an AT-rich region of the cagA promoter that contains an inverted repeat. Variations in the cagA -10 sequence are also observed among the Colombian strains. The location of each motif relative to the cagA transcriptional start site is shown. TS denotes the position of the transcriptional start site. B. Sequences immediately upstream of the cagA ATG initiation site (indicated by a large star) in 8 H. pylori strains were aligned. Four strains express relatively low levels of CagA (5001, 5004, 5005, 5007) and four express relatively high levels of CagA (5043, 5056, 5074, 5093). The AATAAGATA motif is underlined and is present in four strains that express high levels of CagA. The -10 sequences and transcriptional start site (TS) are also shown. C. CagA expression in each strain was analyzed by immunoblotting, and levels of CagA expression were quantified by comparing the CagA signal intensity of each strain to that of a standard (CagA in strain 5001). The relationship between number of AATAAGATA motifs and level of CagA expression is shown. The p-value was determined by t-test.