Abstract
Investigating the evolution of the hepatitis C viral (HCV) genome in the small number of patients that experience viral breakthrough might shed light on the problem of resistance to interferon therapy. Within the HCV genome, sequence diversity of the viral nonstructural 5A protein-coding region (NS5A) has been linked to interferon responsiveness. We analyzed the temporal sequence changes within NS5A in genotype 1a patients: 6 breakthrough (BT), 12 sustained virologic responders (SVR) and 12 non-responders (NR), all of whom had received full dose peg-interferon and ribavirin therapy. The entire NS5A region was amplified by reverse transcription (RT)-PCR followed by direct sequencing of serum samples from baseline and three on-treatment time points for each group. Comparing baseline sequences with week 12 and later time points, BT patients resembled SVR patients in having a higher number of amino acid substitutions at week 12 than NR patients; however, the number of amino acid substitutions in this group decreased at and after BT. Substitutions were focused in the V3 and flanking regions in BT patients but not in SVR patients. The high number of substitutions in NS5A in both BT and SVR groups suggests that selective pressure is associated with viral response to therapy. Our results provide evidence that amino acid substitutions within the NS5A coding region may reflect a host response that drives selective pressure for viral adaptation.
Introduction
During therapy of chronic hepatitis C with peg-interferon (Peg-IFN) and ribavirin (RBV) approximately 54% of patients achieve sustained virologic responses (SVR) (1–3). While most treatment failures are non-responders (NR), who never clear virus from serum, others relapse after completion of therapy. An even smaller group experiences the unique situation of HCV breakthrough (BT), initially responding to therapy, becoming negative for HCV RNA and then developing recurrent viremia despite continued treatment. Initially sensitive to treatment, this group evolves to become resistant, apparently overcoming the effect of interferon on replication. Interferon resistance due to viral or host factors undoubtedly plays a role in each of these scenarios, but the reasons for these response differences are still unknown (4). Response rates within genotypes are fairly consistent for each genotype, implying that a specific viral molecular signature determines responsiveness. While various studies have focused on the envelope protein hypervariable regions (HVRs) and/or the NS5A coding region (5, 6), the locus that defines virologic response differences between genotypes has not been determined. Both E2 and NS5A proteins might interact with IFN-induced cellular protein kinase PKR activity and block its inhibitory activity on the cellular and viral protein synthesis (7, 8). This has not always been supported by other studies (9, 10). In one recent in vitro study, the V3 region and carboxyl terminus of NS5A protein were reported to be responsible for differential IFN resistance (11). The role of NS5A remains controversial; however, it is still a likely target for investigation, given its importance in governing interferon response patterns. We postulated that examining in depth the dynamic molecular changes in NS5A sequentially in patients in three response groups (NR, SVR and BT) at various time points might provide a unique opportunity to explore interferon resistance (12). Previous studies of breakthrough patients have been quite limited, examining only small numbers of patients receiving interferon only or interferon and RBV but not Peg-IFN (13–16). No significant differences in HVR1 and in NS5A were observed when comparing breakthrough or post-breakthrough sequences with baseline suggesting that HCV breakthrough in patients during interferon-based therapy might not be related to selection of an interferon resistant strain. However, it is apparent that even small lapses in compliance can result in breakthrough (17). Since no mention of compliance was made in these studies, it remains unclear whether certain patients might evolve from sensitive to resistant in the face of full compliance with treatment.
The lead-in phase of the Hepatitis C Antiviral Long-term Treatment against Cirrhosis (HALT-C) Trial provided an ideal opportunity to revisit in more detail the evolution of virologic BT (18). We performed a more thorough analysis than previously, by examining temporal sequence changes at intermediate time points during Peg-IFN/RBV therapy, comparing SVR and NR patients with those who experienced BT. Our hypothesis was that changes in NS5A in the early stages of Peg-IFN and RBV treatment would be of particular interest in studying IFN resistance, since observing the molecular evolution from interferon sensitivity to resistance might elucidate the basis for this resistance.
Patients and Methods
Patients and samples
In the lead-in phase of the HALT-C trial, 1,145 patients who had demonstrated previous non-response to interferon-based anti-HCV therapy were treated with Peg-IFN alpha 2a and RBV for 24 weeks; 373 who were HCV RNA negative at week 20 [qualitative Roche COBAS Amplicor™ HCV Test, v. 2.0; lower limit of detection (LOD) 100 IU/mL] (19) were allowed to continue on therapy for a full 48 weeks. Among 43 patients who demonstrated BT, we focused only on the 12 patients who had been fully compliant with therapy (defined as taking 80% of both Peg-IFN and RBV during 80% of the entire treatment period determined by pill counts and detailed interviews at study visit) (20). We assumed that those not receiving the full complement of treatment might experience apparent BT due not to development of de novo interferon resistance but to lack of compliance.
Of the 12 BT patients, three had consecutive HCV RNA positive samples at two time-points, five patients were HCV RNA positive at week 48. The remaining four patients were transiently positive (two at week 24 and two at week 36) but then became negative. All 12 became or remained HCV RNA sero-positive after therapy was discontinued. We chose to focus our analyses on 6 patients infected with HCV genotype 1a, in part because the remaining 6 patients represented different genotypes and also many had low levels of serum HCV RNA at week 12 or the time of breakthrough. Two genotype 3 patients were qualitatively negative at week 12, while the remaining four genotype 1b patients were either qualitatively negative or quantitatively negative; in either case we could not retrieve PCR products for sequencing. Sera from the 6 1a BT patients were available from baseline, week 4, week 12, the time of BT and post BT, all frozen at −80 °C within two hours and shipped to the central NIDDK repository. For comparison, 12 genotype 1a NR patients were matched in blinded fashion by the data-coordinating center for baseline HCV RNA levels, 2:1 with case subjects. Further matching for age, gender or extent of liver disease could not be performed, given the small sample number. Twelve genotype 1a SVR patients who were serum HCV RNA positive by the qualitative Roche COBAS Amplicor™ HCV Test at week 12 were also selected in similar fashion as controls. Samples were used for testing at baseline, week 4, 12 and 24 for NR patients and at baseline, week 4 and 12 for SVR patients. Both NR and SVR patients were also confirmed to have taken at least 80% of both medications. Later samples were not available for NR patients because of their enrollment in the main trial in which patients were randomized 50:50 to maintenance Peg-IFN. The results of the HALT-C Trial have recently been published: no differences in clinical or histological outcomes were observed between the low dose interferon and the control group over 3.8 years (21, 22).
RT-PCR and direct sequencing
HCV RNA was extracted with the QIAamp viral RNA Mini kit (QIAGEN Inc. CA). The HCV NS5A region was amplified as described by Paterson et al (9) with modifications. The first round amplification was performed using the QIAGEN One-step RT-PCR kit (QIAGEN Inc. CA) with outer sense primer (5′-CAGTGGATGAACCGGCTRATA-3′, R=A or G) and antisense primer (5′-TGTGGTGACGTAGCAACGAGTTGCT-3′). The RT-PCR conditions were as follows: 50°C for 30 min, 95°C for 15 min, followed by 35 cycles of 30s at 94°C, 1 min at 49°C and 1.5 min at 72°C with a final extension of 72°C for 10 min. The second round PCR was performed under the same conditions with high fidelity polymerase (Platinum Taq DNA Polymerase High Fidelity, Invitrogen CA) with sense primer (5′-TCCGGTTCCTGGCTAAGRGA-3′) and nested antisense primer (5′-CAGGAGTAAGACATTGAGCAGCAC-3′). The amplification products were purified from positive bands with the QIAquick Gel extraction kit (QIAGEN Inc. CA) and sequenced at the sequencing core facility in UT Southwestern Medical Center with 3 inside forward primers and one reverse primer within NS5A.
Sequence analysis
Sequences of all samples were edited and aligned with Vector NTI software (version 9.0, Invitrogen). Phylogenetic analysis was performed based on the nucleotide sequence using the Neighbor-Joining assay with CLUSTALW embedded in MEGA 4 software. Nucleotide sequences were translated into amino acid sequences. The entire NS5A region was analyzed and the baseline sequences compared with those from later time points, and the number of the deduced amino acid substitutions counted. All sequences analyzed including the baseline sequences and sequences from the later time points have been submitted to GenBank (Accession numbers FJ896264-FJ896370)
The direction of amino acid substitutions in our later time point samples, when compared to baseline, was determined using the NS5A sequence from HCV 1a prototype strain H77 (AF009606) as reference sequence to see whether the amino acid changes at later time points were the same as or different from those in the H77 strain.
Statistical analysis
The Statistical Program for Social Sciences (SPSS 13.0 for windows; SPSS Inc. Chicago, III.) was used for all statistical analyses. The non-parametric test was used to compare the difference between the median numbers of nucleotide and amino acid substitutions in BT patients, NR and SVR patients. The Chi-square test or the Fisher exact test was applied for categorical variables. P values of less than 0.05 were used to indicate statistical significance.
Results
Demographic and virologic characteristics
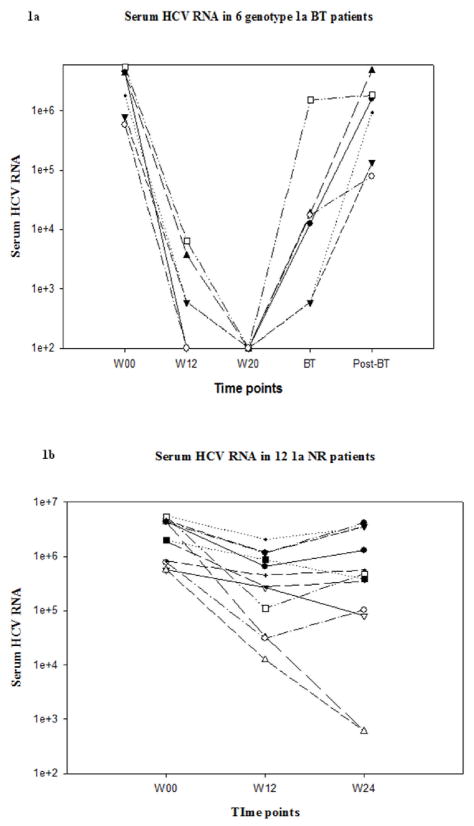
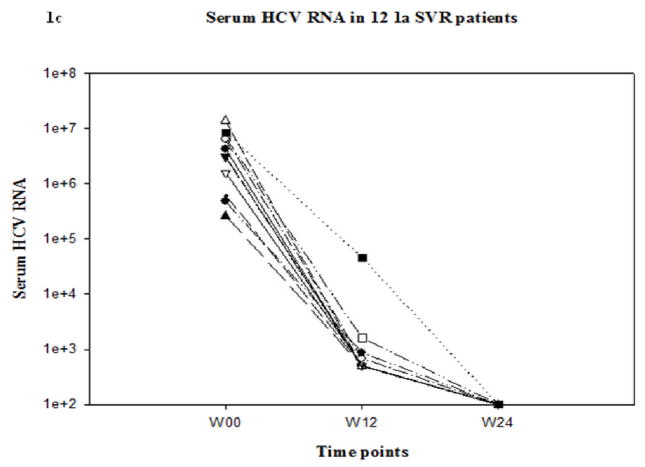
There were no significant differences in the gender, race, age or baseline HCV RNA levels between BT, NR and SVR patients (Table 1). Of the six BT patients, HCV RNA was amplified from all available serum samples at all time points except for two BT patients who were detectable by the qualitative assay but could not be amplified for direct sequencing at week 12. Changes in serum HCV RNA levels from BT, NR and SVR patients are shown in Figure 1. By definition, all BT patients had undetectable serum HCV RNA at week 20 and were continued on full dose therapy. Of the 6 BT patients studied, two demonstrated BT at week 36 and four at week 48 (Figure 1a). Of the 12 SVR patients that had detectable HCV RNA by qualitative PCR at week 12, we could only amplify HCV RNA in 5 patients despite use of multiple primer sets (Figure 1c). The initial decrease in serum HCV viral load was only 0.4 to 2.12 log10 in NR patients. All NR patients had detectable serum HCV RNA levels at all time points (Figure 1b).
Table 1.
Demographic data on all study patients
| Number in each group | NR 12 |
BT 6 |
SVR 12 |
P value |
|---|---|---|---|---|
| Gender (Male/female) | 9/3 | 6/0 | 10/2 | NS |
| Race(Caucasian/Hispanic white/Black) | 10/1/1 | 5/0/1 | 10/2/0 | NS |
| Age (mean ± SD) | 50.6±4.7 | 45.2±5.6 | 47.1±4.9 | NS |
| BMI | 30.7±6.8 | 31.8±3.0 | 30.5±4.6 | NS |
| Baseline viral load (×106 IU/ml) | 3.1 (0.57–5.6) | 3.04 (0.58–5.5) | 3.07 (0.26–14.0) | NS |
BMI= Body mass index
Figure 1.
Serum HCV RNA levels over time in BT(1a), NR(1b) and SVR(1c) patients. Serum HCV RNA levels in all SVR and BT patients were undetectable by definition at week 20. Lower limit of detection was 100 IU/mL.
Phylogenetic analysis
Phylogenetic analysis showed that there were no specific clusters of baseline nucleotide sequences that appear to be related to the different observed responses to Peg-IFN and RBV therapy (Figure 2). Later time point sequences were clustered according to the individual patients, that is, each patient’s sequences more closely resembling other sequences from the same patient, than they did those of other patients.
Figure 2.
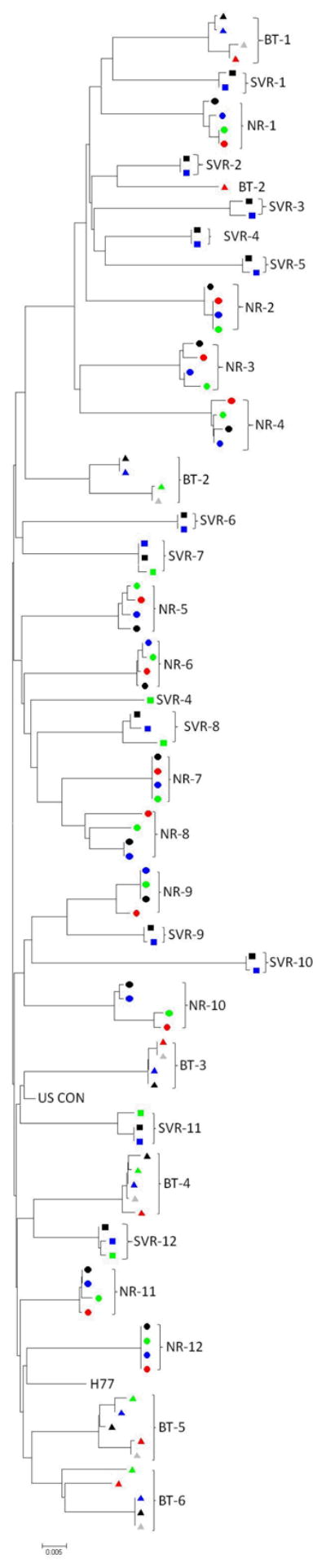
Phylogenetic analysis of sequences from non-responders (●), SVR (■) and BT (▲) patients. In the phylogenetic tree of baseline nucleotide sequences, there were no significant clusters of sequences according to response. When nucleotide sequences from later time points (black: Baseline, blue: wk 4, green: wk 12, red: BT (or wk 24 in NR), grey: post-BT) were added, the later time point sequences diverged slightly from baseline sequences, but still clustered around the patient profile, rather than response to therapy.
Comparison of amino acid substitutions in NS5A region at different time-points between BT, NR and SVR patients
Amino acid substitutions at week 4
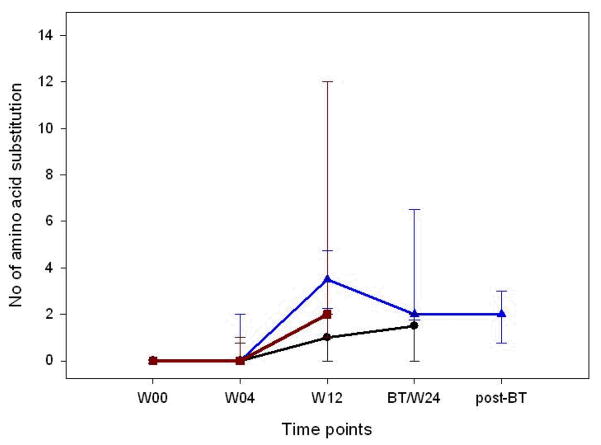
When week 4 sequences were compared with the baseline sequences in each patient, very few amino acid substitutions in NS5A were observed across all treatment response groups despite the presumed drops in viral load in the BT and SVR groups (actual HCV RNA levels at the week 4 time point were not available). Thus, no significant differences in the number of amino acid substitutions were observed between BT, NR and SVR patients (Figure 3: P=NS).
Figure 3.
The number of amino acid substitutions at different time points. The lines represent the median and 25th and 75th percentile value in BT, SVR and NR patients. Blue line represents BT patients; dark red line SVR patients and black line NR patients. BT patients demonstrated the greatest number of amino acid substitutions at week 12, similar to that seen with SVR patients, while NR patients demonstrated little or no genetic drift over time.
Amino acid substitutions at week 12
When week 12 sequences were compared with baseline sequences for each patient, all patient groups had more amino acid substitutions at week 12 compared to week 4 [Figure 3, BT patients (n=4): P=0.10; SVR patients (n=5): P= 0.06; NR patients (n=12): P=0.014, respectively]. However, the median number of amino acid substitutions at week 12 was greater in BT (3, range 2–4) and SVR (2, range 1–22) than in NR patients (1, range 0–5, P=0.045 respectively for trend). In addition, BT patients had a significantly higher median number of amino acid substitutions at week 12 compared to those observed in NR patients (Figure 3, P=0.02).
Amino acid substitutions at the time of viral breakthrough in BT patients and week 24 in NR patients
When the sequences at the time of BT and the week 24 sequences from NR patients were compared with their baseline sequences, the number of amino acid substitutions appeared to decline in BT patients when compared with the number of amino acid substitutions at week 12. Very few changes in the number of amino acid substitutions were evident in NR patients after week 12 (Figure 3).
Thus BT patients and NR patients had comparable median numbers of amino acid substitutions in the later stages of treatment [after week 12, BT: 2 (1–20), vs. NR: 1.5 (0–7), P=0.25]. Because all BTs occurred after week 24 and later time points were not available for NR patients because they were enrolled at week 24 in the long-term study, the error rate of amino acid substitutions per site per year was calculated to eliminate the effect of the difference in the duration between the BT patients and NR patients (NR measured at week 24). Using this correction, we still found no significant difference in the error rate of amino acid substitutions between BT patients and NR patients [BT: 0.005 (0.002–0.064), vs. NR: 0.007 (0–0.034), P=0.96].
Amino acid substitutions in the individual regions within NS5A protein at week 12
To study changes in the distinct regions within NS5A, we initially analyzed the amino terminal and carboxyl terminal regions, comparing aa1-236 to aa237-448. In the amino terminus, no significant difference was found in the median number of amino acid substitutions among BT, SVR and NR patients. However, in the carboxyl terminus, BT and SVR patients demonstrated a higher median number of amino acid substitutions compared with NR patients. [SVR: 2 (0–12); BT: 2 (1–5); NR: 0 (0–2) P= 0.055 for trend].
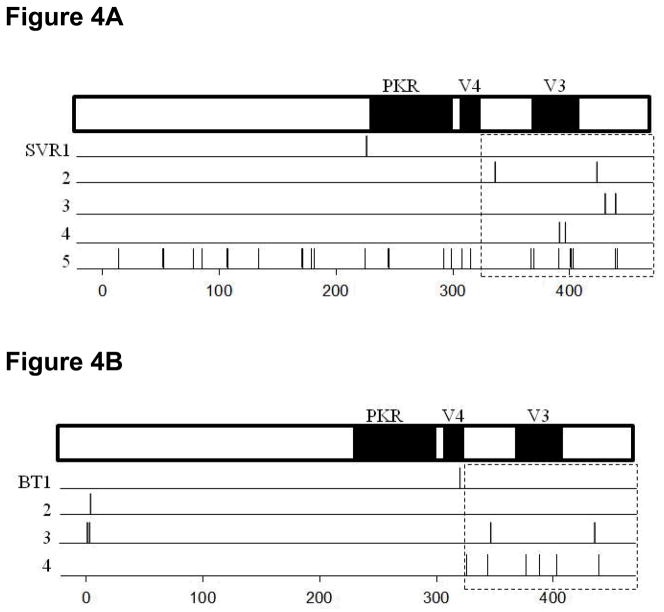
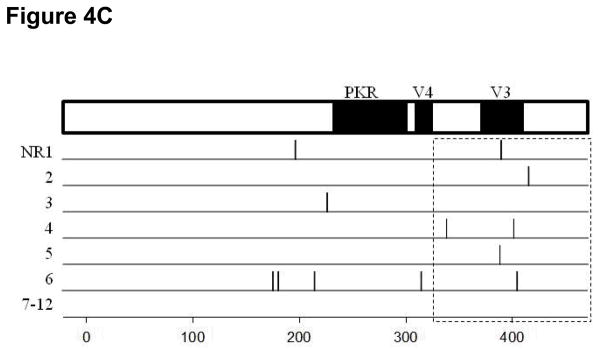
When the carboxyl terminal region was further divided into the small hot-spot regions already documented to be associated with differential responses in published papers (such as PKR, V3, V4 regions) (5, 23, 24), only the region downstream of V3 showed a significantly higher median number of amino acid substitutions in BT and SVR patients compared with NR patients (Figure 4, P=0.022). The region upstream of V3 tended to have a higher median number of amino acid substitutions in BT patients compared with NR patients but this was not significant (Figure 4B and C, P=0.078). No significant differences were found within the PKR, V4 and V3 regions. We did not identify any specific amino acid substitutions that were consistently present which would point to a strong correlation with specific responses (BT, SVR or NR) to therapy. We did not observe any amino acid substitutions in the carboxyl terminal (AA350-356) proline-rich region (25, 26) or in the putative hyperphosphorylation sites (S222, S225, S229 and S232) (27).
Figure 4.
The distribution of amino acid substitutions across the NS5A region in the period between week 4 and week 12 in SVR (Panel A), BT (Panel B) and NR (Panel C) patients. Each line represents one patient, with vertical lines indicating amino acid substitutions between weeks 4 and 12. Most substitutions occurred in the carboxyl terminal region around V3. There were more amino acid substitutions overall in the SVR and BT patients than were observed in the NR patients. Six NR patients, listed as 7–12, demonstrated no mutations whatsoever in this region.
Direction of amino acid substitutions in the NS5A region
While SVR and BT patients had comparable numbers of nucleotide and amino acid substitutions at week 12, they experienced different final outcomes with Peg-IFN and RBV therapy. We therefore examined the direction of amino acid substitutions as compared to the HCV 1a prototype sequence H77. We made the comparison between week 4 and week 12, since most of the amino acid substitutions occurred during this period. Of interest, no amino acid substitutions reverted toward baseline between week 4 and week 12.
In five SVR patients who had NS5A sequence data available at week 12, high degree of variability in the number of amino acid substitutions between SVR patients was observed. A total of 29 amino acid substitutions were found (mean number/patient 5.8). Among these five SVR patients, a total of 14 (range 1–8) amino acids were different from those in the HCV 1a prototype sequence H77. Only two of five SVR patients showed (1 and 14) amino acid substitutions that were identical to those in the H77 strain. These amino acid substitutions were evenly distributed across the entire NS5A region. Around 45% (13/29) of amino acid substitutions were located in the carboxyl terminal regions: three upstream of the V3 region (aa331-380), five in the V3 region and five downstream of V3 (aa441-448) (Figure 4A).
In the four BT patients who could be sequenced for NS5A, a total of 12 amino acid substitutions were found from week four to week 12 (mean number 3/patient). Three of four BT patients showed one, three or four amino acid substitutions respectively that became different with those in the H77 strain. All of these mutations were transient and were not observed at later time points. At the time of BT, 7 of 8 amino acid substitutions had reverted to the baseline sequence, while the remaining one matched that in the H77 strain. In the same time frame (week four to week 12), three of four BT patients each showed a total of 4 amino acid substitutions toward the H77 strain. Three of the four amino acid substitutions remained unchanged for the duration of treatment. One amino acid substitution was different from that in the H77 strain and baseline strain by the time of viral breakthrough. Overall, the majority of these mutations (8/12, 67%) were located in the carboxyl terminal region: 4 upstream of the V3 region (aa331-380), 2 in the V3 region itself and 2 downstream of V3 (aa441-448) (Figure 4B).
Among the 12 NR patients, only 12 amino acid substitutions were observed from week 4 to week 12 across all of NS5A (mean number 1/patient). Six amino acid substitutions were different from those in the H77 strain. Of these, five mutated back to baseline ones between week 12 and 24, and the remaining amino acid substitutions remained unchanged during this period. The other 6 amino acid substitutions that became same as those in the H77 stain at week 12 remained stable between week 12 and 24. Half (6/12) of the amino acid substitutions were located in the carboxyl terminal region: 1 upstream of the V3 region (aa331-380), 4 in the V3 and 1 downstream of the V3 region (aa441-448) (Figure 4C).
Discussion
Previous studies have shown that certain HCV genotypes and low HCV RNA levels at baseline are associated with increased likelihood of sustained viral response to interferon therapy (28). Few studies have looked at changes in the HCV genomic sequence over time and whether the emergence of specific sequences might be associated with non-response (5, 29). Our principal finding was that BT patients, like those who would later experience SVR, demonstrated a larger number of amino acid substitutions occurring between week 4 and week 12 than did NR patients. However, once breakthrough occurred, the pattern of amino acid substitutions in BT patients did not persist but reverted toward baseline, with reversal or negation of virtually all previous mutations.
We found an insignificant number of amino acid substitutions in the NS5A region within the first four weeks of therapy in any of the three groups. Other studies have shown few changes in consensus sequences during this period (30, 31). Examining consensus sequences only, however, is likely to be less sensitive than quasi-species analysis in determining changes in overall molecular patterns; these studies are planned. The higher number of amino acid substitutions observed in BT and SVR but not in NR patients between weeks 4 and 12 implies that divergence of HCV occurred primarily during the period of low HCV replication (or was more evident when HCV RNA had nearly reached its nadir). In this phase, an enhanced immune response or the cumulative effects of RBV may be more capable of driving the change in HCV quasi-species that results in more sequence changes (16). Immune effects may include clearance of IFN sensitive variants, accumulation of existing IFN-resistant fit sequences or development of de novo IFN-resistant mutations (32). An alternative explanation might be that the evolution of the dominant variants that will eventually become the consensus sequence is likely to be more readily evident when the viral load is lowest.
By contrast, NR patients demonstrated little or no substitutions in NS5A during therapy, despite viral load declines of 0.4–2.12 logs at week 12, suggesting that the virus in NR patients is intrinsically highly fit and resistant to the antiviral effect of interferon (5, 7). Alternatively, intrinsic or virus-directed defects of the innate and/or adaptive immune responses may be responsible for this apparent absence of interferon pressure (33). By contrast, SVR patients showed many more amino acid substitutions from week four to week 12, evenly distributed throughout the entire NS5A region, suggesting that targets of selective pressure in SVR patients are widely distributed with no specific hot spots identified. Despite this high mutation rate, viral suppression continues, eventually resulting in SVR. Our single patient with high mutation number suggests that there may be great variability in immune pressure in certain instances or at certain times, but further study will be necessary to confirm this finding.
Like the SVR patients, BT patients also showed a higher number of amino acid substitutions between weeks 4 and 12, suggesting that BT patients also experience enhanced selective pressure during the low replication phase. Unlike the SVR patients, 67% of the amino acid substitutions observed in BT patients focused on the carboxyl terminal region, specifically V3 and its flanking regions. This might indicate a role for V3 in modulating the intracellular antiviral activity leading to the subsequent breakthrough event (23, 34). On the other hand, three of the four BT patients had amino acid substitutions which came to resemble more closely the HCV 1a prototype sequence H77 between week 4 and week 12, suggesting either that selective pressure in BT patients might lead to creation of this apparently highly fit clone or that a defective immune response develops in BT as compared to SVR patients who have an ongoing clearance response. Finally, the reversion of previously changed amino acids toward the patient’s own baseline sequence after viral breakthrough would suggest that the immune dysfunction and/or loss of selective pressure in the BT patients occurs in this late phase of antiviral therapy. An initially strong immune response might lead to suppression of the dominant fit virus in favor of minor variants that later, when immune pressure becomes weaker, allow the original strain to re-emerge. It was not possible in this study to determine whether the defect in the selective pressure was due to viral subversion of the intracellular antiviral mechanism (33) or another mechanism such as a down-regulation of IFN receptors (35, 36) or even a loss of appropriate cellular immune responses. We excluded patients who had demonstrated non-compliance; all patients had evidence of at least 80% compliance with both medications. Other reasons for BT could include unreported or unrecognized non-compliance.
NS5A has been ascribed as having multiple functions in viral replication and host cell regulation, and specific mutations in NS5A have been shown to positively or negatively impact viral RNA replication (37). The distinct amino and carboxyl terminal regions of NS5A have been respectively implicated in sub-cellular localization and protein interactions, wherein the sequence changes in the former have been shown to negatively impact viral replication (38, 39). However, sequence alterations and insertions in the carboxyl terminal region of NS5A are often well tolerated (37, 40). Our study suggests that NS5A amino acid substitutions differing from the H77 sequence were largely clustered within the carboxyl terminal region of the protein from SVR and BT patients. This is consistent with previous observations of NS5A carboxyl-terminal region sequence diversity among HCV patients (37), and suggests that the NS5A carboxyl terminus is a genetically flexible domain. Changes in the consensus sequence or in the complement of minor IFN-resistant viruses might be capable of subverting the intracellular antiviral activity and result in infected cells becoming more suitable for HCV replication (33). It is also possible that genetic flexibility is accompanied by compensatory changes elsewhere in the viral genome, such as in E2, and that these compensatory changes affect overall viral fitness and response to IFN therapy (41). Further studies will focus on quasi-species analysis using cloning techniques to address these questions.
In conclusion, although few mutations appeared within the first four weeks of treatment, patients responding to treatment demonstrated more mutations in the NS5A coding region than non-responders by week 12. Initially, SVR and BT patients demonstrated similar numbers of nucleotide and amino acid substitutions. Subsequently, SVR patients became virus-negative while BT patients, initially virus-negative, eventually reverted to baseline at the time of BT and thereafter. Of note, the NS5A amino acid substitutions in BT patients focused in V3 and flanking regions but there was not one specific mutation that characterized this group. These results suggest that the enhanced selective pressure is vigorous and multi-specific in SVR patients while being ineffective or absent in non-responders. BT patients develop an apparent defect in immune pressure during treatment that results in no further mutations and, in fact, reversion towards their baseline configuration. We hypothesize that specific viral quasi-species could impart dominant effects on the virus/host processes that direct the interferon response and control of HCV replication.
Acknowledgments
ACKNOWLEDGMENTS/FUNDING SOURCES
This study was supported by the National Institute of Diabetes & Digestive & Kidney Diseases (contract numbers are listed below). Additional support was provided by the National Institute of Allergy and Infectious Diseases (NIAID), the National Cancer Institute, the National Center for Minority Health and Health Disparities and by General Clinical Research Center grants from the National Center for Research Resources, National Institutes of Health (grant numbers are listed below). Additional funding to conduct this study was supplied by Hoffmann-La Roche, Inc., through a Cooperative Research and Development Agreement (CRADA) with the National Institutes of Health.
In addition to the authors of this manuscript, the following individuals were instrumental in the planning or conduct of this study at each of the participating institutions as follows:
University of Texas Southwestern Medical Center, Dallas, TX: (Contract N01-DK-9-2321, Grant M01RR-00633) Thomas E. Rogers, MD, Janel Shelton, Nicole Crowder, LVN, Rivka Elbein, RN, BSN
University of Washington, Seattle, WA: (Contract N01-DK-9-2318) Stacy Homer, Ph.D.
New England Research Institutes, Watertown, MA: (Contract N01-DK-9-2328) Margaret C. Bell, MS, MPH, Linda Massey
National Institute of Diabetes and Digestive and Kidney Diseases, Division of Digestive Diseases and Nutrition, Bethesda, MD: James E. Everhart, MD, Leonard B. Seeff, MD, Patricia R. Robuck, PhD, Jay H. Hoofnagle, MD, Elizabeth C. Wright, PhD
Data and Safety Monitoring Board Members: (Chair) Gary L. Davis, MD, Guadalupe Garcia-Tsao, MD, Michael Kutner, PhD, Stanley M. Lemon, MD, Robert P. Perrillo, MD
Footnotes
This is publication #35 of the HALT-C Trial
Financial Disclosures Paragraph for Insertion into Manuscript
Financial relationships of the authors with Hoffmann-La Roche, Inc., are as follows: W.M. Lee and M. Jain receive research support. Authors with no financial relationships related to this project are: H.-J. Yuan, K.K. Snow, and M. Gale, Jr.
References
- 1.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347(13):975–82. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 2.Hadziyannis SJ, Sette H, Jr, Morgan TR, et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140(5):346–55. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- 3.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358(9286):958–65. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 4.Yuan HJ, Lee WM. Nonresponse to treatment for hepatitis C: current management strategies. Drugs. 2008;68(1):27–42. doi: 10.2165/00003495-200868010-00003. [DOI] [PubMed] [Google Scholar]
- 5.Enomoto N, Sakuma I, Asahina Y, et al. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334(2):77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 6.Farci P, Shimoda A, Coiana A, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288(5464):339–44. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 7.Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285(5424):107–10. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 8.Gale M, Jr, Blakely CM, Kwieciszewski B, et al. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18(9):5208–18. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paterson M, Laxton CD, Thomas HC, Ackrill AM, Foster GR. Hepatitis C virus NS5A protein inhibits interferon antiviral activity, but the effects do not correlate with clinical response. Gastroenterology. 1999;117(5):1187–97. doi: 10.1016/s0016-5085(99)70405-1. [DOI] [PubMed] [Google Scholar]
- 10.Podevin P, Sabile A, Gajardo R, et al. Expression of hepatitis C virus NS5A natural mutants in a hepatocytic cell line inhibits the antiviral effect of interferon in a PKR-independent manner. Hepatology. 2001;33(6):1503–11. doi: 10.1053/jhep.2001.24749. [DOI] [PubMed] [Google Scholar]
- 11.Tsai YH, Kuang WF, Lu TY, et al. The non-structural 5A protein of hepatitis C virus exhibits genotypic differences in interferon antagonism. J Hepatol. 2008;49(6):899–907. doi: 10.1016/j.jhep.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 12.Germanidis G, Metallidis S, Lazaraki G, Pawlotsky JM, Nikolaidis P. NS5A sequences of hepatitis C virus genotype 1 and interferon resistance: where are we? J Infect Dis. 2008;198(1):154–5. doi: 10.1086/588709. [DOI] [PubMed] [Google Scholar]
- 13.Vuillermoz I, Khattab E, Sablon E, et al. Genetic variability of hepatitis C virus in chronically infected patients with viral breakthrough during interferon-ribavirin therapy. J Med Virol. 2004;74(1):41–53. doi: 10.1002/jmv.20144. [DOI] [PubMed] [Google Scholar]
- 14.Polyak SJ, McArdle S, Liu SL, et al. Evolution of hepatitis C virus quasispecies in hypervariable region 1 and the putative interferon sensitivity-determining region during interferon therapy and natural infection. J Virol. 1998;72(5):4288–96. doi: 10.1128/jvi.72.5.4288-4296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeuzem S, Lee JH, Roth WK. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25(3):740–4. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]
- 16.Farci P, Strazzera R, Alter HJ, et al. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc Natl Acad Sci U S A. 2002;99(5):3081–6. doi: 10.1073/pnas.052712599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiffman ML, Ghany MG, Morgan TR, et al. Impact of reducing peginterferon alfa-2a and ribavirin dose during retreatment in patients with chronic hepatitis C. Gastroenterology. 2007;132(1):103–12. doi: 10.1053/j.gastro.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Lee WM, Dienstag JL, Lindsay KL, et al. Evolution of the HALT-C Trial: pegylated interferon as maintenance therapy for chronic hepatitis C in previous interferon nonresponders. Control Clin Trials. 2004;25(5):472–92. doi: 10.1016/j.cct.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Morishima C, Morgan TR, Everhart JE, et al. HCV RNA detection by TMA during the hepatitis C antiviral long-term treatment against cirrhosis (Halt-C) trial. Hepatology. 2006;44(2):360–7. doi: 10.1002/hep.21265. [DOI] [PubMed] [Google Scholar]
- 20.McHutchison JG, Manns M, Patel K, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology. 2002;123(4):1061–9. doi: 10.1053/gast.2002.35950. [DOI] [PubMed] [Google Scholar]
- 21.Di Bisceglie AM, Shiffman ML, Everson GT, et al. Prolonged therapy of advanced chronic hepatitis C with low-dose peginterferon. N Engl J Med. 2008;359(23):2429–41. doi: 10.1056/NEJMoa0707615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiffman ML, Di Bisceglie AM, Lindsay KL, et al. Peginterferon alfa-2a and ribavirin in patients with chronic hepatitis C who have failed prior treatment. Gastroenterology. 2004;126(4):1015–23. doi: 10.1053/j.gastro.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Nousbaum J, Polyak SJ, Ray SC, et al. Prospective characterization of full-length hepatitis C virus NS5A quasispecies during induction and combination antiviral therapy. J Virol. 2000;74(19):9028–38. doi: 10.1128/jvi.74.19.9028-9038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy MD, Rosen HR, Marousek GI, Chou S. Analysis of sequence configurations of the ISDR, PKR-binding domain, and V3 region as predictors of response to induction interferon-alpha and ribavirin therapy in chronic hepatitis C infection. Dig Dis Sci. 2002;47(6):1195–205. doi: 10.1023/a:1015349924116. [DOI] [PubMed] [Google Scholar]
- 25.Tan SL, Nakao H, He Y, et al. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci U S A. 1999;96(10):5533–8. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y, Nakao H, Tan SL, et al. Subversion of cell signaling pathways by hepatitis C virus nonstructural 5A protein via interaction with Grb2 and P85 phosphatidylinositol 3-kinase. J Virol. 2002;76(18):9207–17. doi: 10.1128/JVI.76.18.9207-9217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan SL, Katze MG. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology. 2001;284(1):1–12. doi: 10.1006/viro.2001.0885. [DOI] [PubMed] [Google Scholar]
- 28.Berg T, Sarrazin C, Herrmann E, et al. Prediction of treatment outcome in patients with chronic hepatitis C: significance of baseline parameters and viral dynamics during therapy. Hepatology. 2003;37(3):600–9. doi: 10.1053/jhep.2003.50106. [DOI] [PubMed] [Google Scholar]
- 29.Donlin MJ, Cannon NA, Yao E, et al. Pretreatment sequence diversity differences in the full-length hepatitis C virus open reading frame correlate with early response to therapy. J Virol. 2007;81(15):8211–24. doi: 10.1128/JVI.00487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulestin A, Sandres-Saune K, Payen JL, Rostaing L, Pasquier C, Izopet J. Genetic heterogeneity of the NS5A gene of hepatitis C virus and early response to interferon-alpha. J Infect Dis. 2003;188(9):1367–70. doi: 10.1086/379037. [DOI] [PubMed] [Google Scholar]
- 31.Dal Pero F, Tang KH, Gerotto M, et al. Impact of NS5A sequences of Hepatitis C virus genotype 1a on early viral kinetics during treatment with peginterferon- alpha 2a plus ribavirin. J Infect Dis. 2007;196(7):998–1005. doi: 10.1086/521306. [DOI] [PubMed] [Google Scholar]
- 32.Bowen DG, Walker CM. The origin of quasispecies: cause or consequence of chronic hepatitis C viral infection? J Hepatol. 2005;42(3):408–17. doi: 10.1016/j.jhep.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Gale M, Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436(7053):939–45. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- 34.Duverlie G, Khorsi H, Castelain S, et al. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J Gen Virol. 1998;79 ( Pt 6):1373–81. doi: 10.1099/0022-1317-79-6-1373. [DOI] [PubMed] [Google Scholar]
- 35.Fukuda R, Ishimura N, Hamamoto S, et al. Co-infection by serologically-silent hepatitis B virus may contribute to poor interferon response in patients with chronic hepatitis C by down-regulation of type-I interferon receptor gene expression in the liver. J Med Virol. 2001;63(3):220–7. doi: 10.1002/1096-9071(200103)63:3<220::aid-jmv1004>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Mizukoshi E, Kaneko S, Yanagi M, et al. Expression of interferon alpha/beta receptor in the liver of chronic hepatitis C patients. J Med Virol. 1998;56(3):217–23. doi: 10.1002/(sici)1096-9071(199811)56:3<217::aid-jmv7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 37.Macdonald A, Harris M. Hepatitis C virus NS5A: tales of a promiscuous protein. J Gen Virol. 2004;85(Pt 9):2485–502. doi: 10.1099/vir.0.80204-0. [DOI] [PubMed] [Google Scholar]
- 38.Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J Virol. 2008;82(3):1073–83. doi: 10.1128/JVI.00328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brass V, Pal Z, Sapay N, et al. Conserved determinants for membrane association of nonstructural protein 5A from hepatitis C virus and related viruses. J Virol. 2007;81(6):2745–57. doi: 10.1128/JVI.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moradpour D, Evans MJ, Gosert R, et al. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J Virol. 2004;78(14):7400–9. doi: 10.1128/JVI.78.14.7400-7409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Z, Fan X, Xu Y, Di Bisceglie AM. Comparative analysis of nearly full-length hepatitis C virus quasispecies from patients experiencing viral breakthrough during antiviral therapy: clustered mutations in three functional genes, E2, NS2, and NS5a. J Virol. 2008;82(19):9417–24. doi: 10.1128/JVI.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]