Figure 8.
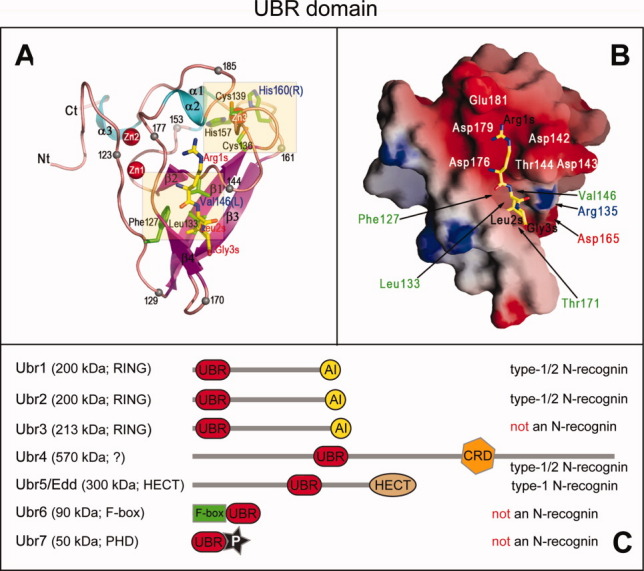
UBR domains in N-recognins and putative N-recognins of the mammalian Arg/N-end rule pathway. A: Ribbon diagram of the ∼80-residue S. cerevisiae UBR domain83 (Fig. 7A) in the complex with the RLGES peptide that bears N-terminal Arg, a Type-1 Ndp residue.28 The bound RLGES is shown as a stick model, with carbon atoms colored yellow. Several residues are marked with a black sphere and numbered to facilitate the tracing of the polypeptide chain. The names of residues of the RLGES peptide are in red, with the letter “s” (substrate) appended to their position numbers. Side chains of residues in the UBR domain that are present near missense mutations in UBR1 of patients with Johanson–Blizzard syndrome (JBS; C.-S. Hwang et al., unpublished data) are shown in a stick form, with carbon atoms colored green. Three coordinated zinc ions of the UBR domain83 are shown as red spheres. B: Molecular surface of the S. cerevisiae UBR domain. Negatively and positively charged surfaces are shaded red and blue, respectively. The bound RLGES peptide is shown in yellow. Some residues of Ubr1 that comprise the N-degron-binding cleft are labeled.83 C: Diagram of the mammalian UBR-domain family of E3 Ub ligases, showing both UBR and other domains of these E3s (RING, HECT, PHD, CRD and F-box) that contribute to recognition and ubiquitylation of protein substrates.31,43 Ubr1, Ubr2, Ubr4, and Ubr5/Edd of this set are operationally defined N-recognins of the mammalian Arg/N-end rule pathway in that they specifically bind to the Type-1 and/or Type-2 destabilizing N-terminal residues, whereas Ubr3, Ubr6, and Ubr7 are not N-recognins43,274 (The double-E3 design of the Arg/N-end rule pathway section). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
