Figure 11.
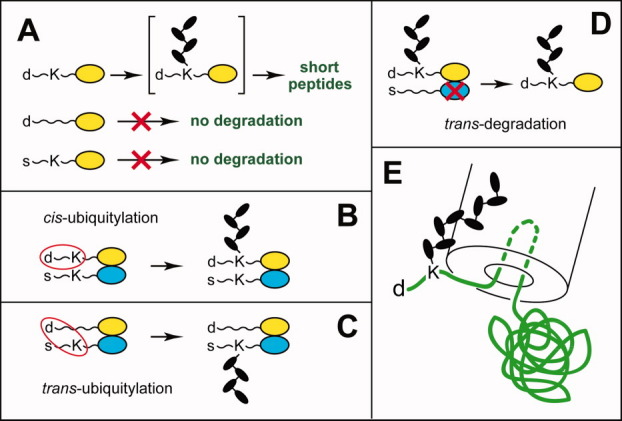
Organization and cis-trans targeting of eukaryotic N-degrons. A: Three determinants of N-degron. d, a destabilizing N-terminal residue. K, a “ubiquitylatable” internal Lys residue. The absence of one of these determinants abrogates polyubiquitylation of a protein, despite the presence of another determinant. The third determinant of N-degron is an unstructured region that is required for polyubiquitylation and/or the initiation of degradation of a polyubiquitylated N-end rule substrate by the 26S proteasome. See Substrates of the N-End Rule Pathway section for references and details. B and C: cis versus trans polyubiquitylation of an oligomeric N-end rule substrate that results in the degradation of a subunit that becomes linked to a poly-Ub chain13 (see Substrates of the N-End Rule Pathway section). D: trans-degradation, in which a specific subunit of oligomeric protein is polyubiquitylated but is not degraded by the 26 proteasome, for example, because it lacks an unstructured region that is required for the initiation of degradation. Instead, a subunit-selective degradation of another, nonubiquitylated subunit takes place. This mode of degradation was demonstrated by the Matouschek laboratory168 for oligomeric substrates of the UFD pathway. It remains to be determined whether the analogous (hypothetical) trans-degradation of an oligomeric N-end rule substrate can also occur. E: The 1989–1996 hairpin insertion model of protein targeting by the 26S proteasome.5 No details of the 26S proteasome structure (such as the 19S regulatory particle (RP)) are shown in this 1996 diagram,5 and the sizes of specific components such as Ub moieties, the poly-Ub chain and the proteasome, are not to scale (see Substrates of the N-End Rule Pathway section). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
