Abstract
Desiccation tolerance, the ability to survive nearly total dehydration, is a rare strategy for survival and reproduction observed in all taxa. However, the mechanism and regulation of this phenomenon are poorly understood. Correlations between desiccation tolerance and potential effectors have been reported in many species, but their physiological significance has not been established in vivo. Although the budding yeast Saccharomyces cerevisiae exhibits extreme desiccation tolerance, its usefulness has been hampered by an inability to reduce tolerance more than a few fold by physiological or genetic perturbations. Here we report that fewer than one in a million yeast cells from low-density logarithmic cultures survive desiccation, while 20–40% of cells from saturated cultures survive. Using this greatly expanded metric, we show that mutants defective in trehalose biosynthesis, hydrophilins, responses to hyperosmolarity, and hypersalinity, reactive oxygen species (ROS) scavenging and DNA damage repair nevertheless retain wild-type levels of desiccation tolerance, suggesting that this trait involves a unique constellation of stress factors. A genome-wide screen for mutants that render stationary cells as sensitive as log phase cells identifies only mutations that block respiration. Respiration as a prerequisite for acquiring desiccation tolerance is corroborated by respiration inhibition and by growth on nonfermentable carbon sources. Suppressors bypassing the respiration requirement for desiccation tolerance reveal at least two pathways, one of which, involving the Mediator transcription complex, is associated with the shift from fermentative to respiratory metabolism. Further study of these regulators and their targets should provide important clues to the sensors and effectors of desiccation tolerance.
DESICCATION tolerance is the ability of an organism to withstand removal of its intracellular liquid water and then resume normal metabolism after rehydration (Crowe et al. 1992). This trait is common in the seeds of most gametophytes, which contain desiccated plant embryos (Finkelstein et al. 2008). However, rare desiccation tolerant “adult” species exist such as baker’s yeast (Lesaffre Yeast Corporation 2010), resurrection plants (Bartels 2005), and tardigrades (Sømme 1996), which are broadly distributed among the taxa of unicellular microbes and multicellular plants and animals, respectively. The ability of seeds and these extremophiles to survive desiccation is thought to depend upon their unique ability to mitigate the deleterious consequences of possibly multiple stresses occurring at all scales, from biochemical reactions to interactions among tissues (Tunnacliffe and Ricci 2006). However, the molecular basis of desiccation tolerance remains unknown. Indeed, fundamental questions remain unanswered. What are the stresses imposed by desiccation? Which of these stresses are lethal to sensitive organisms? What differences in the physiology or stress responses of tolerant organisms allows them to prevent or overcome the damage imposed by desiccation? How is desiccation tolerance regulated? Addressing these questions will contribute to the understanding of this remarkable trait and potentially reveal basic insights into water homeostasis in all cells.
The impact of desiccation on cells can potentially be understood as a composite of several component stresses. Evaporation of water from the medium surrounding a cell increases external solute concentrations, leading to hyperosmotic stress and hypersalinity. Unmitigated, these stresses would lead to increased concentrations of intracellular macromolecules and ions potentially generating cellular toxicity through aggregation, altered reaction kinetics, or production of toxins such as reactive oxygen species (ROS) (Jiang and Zhang 2002; Kranner and Birtić 2005). These toxic molecules may in turn damage DNA, proteins, or membranes. As a result, a number of stress-response pathways and molecules have been hypothesized to be important for desiccation tolerance, including osmoregulation, ion homeostasis, DNA damage repair, and protein folding.
Indeed, several potential stress-response molecules and pathways appear to be induced in the desiccation-tolerant state of desiccation-tolerant seeds and extremophiles (Goyal et al. 2005). One class of induced molecules consists of nonreducing disaccharides such as trehalose and sucrose. These compounds are thought to act as compatible solutes, maintaining osmotic balance during moderate water deficit, and perhaps exhibiting protective effects as water removal becomes extreme. Another class of induced molecules is a family of inherently unstructured hydrophilic proteins known as hydrophilins or late embryogenesis abundant (LEA) proteins (Garay-Arroyo et al. 2000; Battaglia et al. 2008). Some hydrophilins are proposed to stabilize membranes and native protein structures, either alone or in concert with trehalose (Sales et al. 2000). In addition to these small molecules, proteins dedicated to stress responses are also induced. For example, heat shock proteins that correct protein misfolding are induced during seed desiccation (Almoguera and Jordano 1992; Wehmeyer et al. 1996). Desiccation also triggers the conserved high osmolarity glycerol (HOG) pathway in yeast (O’Rourke et al. 2002) and other species (Al-Rubeai et al. 2007).
However, the functional links between desiccation tolerance and these correlated stress-response pathways and molecules are either absent or controversial. For example, desiccation tolerance in the budding yeast Saccharomyces cerevisiae was interpreted to be dependent on trehalose accumulation in one study (Gadd et al. 1987), but independent of it in another (Ratnakumar and Tunnacliffe 2006). Specific genetic tests for the role of other candidate effectors such as hydrophilins or stress-response regulators of osmolarity, DNA damage repair, ROS, or salinity have not been performed. Therefore, it is unclear what stresses cause desiccation sensitivity and what stress responses are necessary for desiccation tolerance. Many of the stress factors correlated with desiccation are induced in response to most other stresses as well (Gasch et al. 2000), suggesting that desiccation stress factors may be neither unique nor novel. Furthermore, while desiccation tolerant organisms appear to exist in both desiccation sensitive and tolerant states (Crowe 1972; Gadd et al. 1987; Wright 1988), how they regulate these states is unknown. One clue may come from adult plants for which water reduction leads to a transcriptional response initiated by the phytohormone abscisic acid (Zhu 2002). However, how this response to mild water reduction relates to true desiccation tolerance is unknown.
To address many of these fundamental questions about desiccation tolerance, the budding yeast S. cerevisiae appears to be an ideal model organism. In its desiccation-tolerant state it shares many of the correlates of other desiccation-tolerant species or tissues such as the increased presence of trehalose and hydrophilins (Gadd et al. 1987; Sales et al. 2000; Singh et al. 2005). Furthermore, this organism is used intensively to study the deleterious effects of a number of stresses like heat shock, high osmolarity, and DNA damage as well as the highly conserved pathways that respond to those stresses. However, to date the use of these potential attributes of yeast have been limited. No systematic study of the role of known stress responses in desiccation tolerance has been reported. A few studies have performed unbiased screens for reduced desiccation tolerance of deletion mutants of nonessential genes of yeast (D’Elia et al. 2005; Shima et al. 2008). These studies have identified many different mutants that reduce the survival of desiccated cells by up to 20-fold. While potentially informative, the relatively small effect on viability makes further phenotypic and genetic studies of these mutants challenging.
Here our studies of desiccation tolerance of wild-type yeast reveal that exponential and saturated yeast cultures can exhibit as much as a one-million-fold difference in desiccation tolerance. Using this more robust metric, we systematically assess the causal role of stress-response pathways in desiccation tolerance and perform unbiased screens for mutants that promote desiccation sensitivity or tolerance. Our results establish budding yeast as a powerful system to elucidate the molecular basis of desiccation tolerance.
Materials and Methods
Yeast strains
Standard methods were used for strain construction (Table 1) (Rose et al. 1990).
Table 1 . Saccharomyces cerevisiae and other strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| BY4742 | MATα his3∆1 leu2∆0 lys2∆0 ura3∆0 | ATCC |
| cat1∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 cat1::KanR | ATCC |
| cta1∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 cta1::KanR | ATCC |
| gre1∆ sip18∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 gre1::KanR sip18::KanR | This study |
| gre1∆ hsp12∆ sip18∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 gre1::KanR sip18::KanR hsp12::HygR | This study |
| gre1∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 gre1::KanR | ATCC |
| hal1∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 hal12::KanR | ATCC |
| hal5∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 hal5::KanR | ATCC |
| hsp12∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 hsp12::KanR | ATCC |
| hsp104∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 hsp104::KanR | ATCC |
| hog1∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 hog1::KanR | ATCC |
| met22∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 met22::KanR | ATCC |
| mrpl16∆ | MATα his3∆1 leu2∆0 lys2∆0 ura3∆0 mrpl16::KanR | |
| msb2∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 msb2::KanR | ATCC |
| msn2∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 msn2::KanR | ATCC |
| msn4∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 msn4::KanR | ATCC |
| msn2∆ msn4∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 msn2::KanR msn4::HygR | This study |
| nst1∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 nst1::KanR | ATCC |
| opy2∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 opy2::KanR | ATCC |
| pbs2∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 pbs2::KanR | ATCC |
| rad52∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 rad52::KanR | ATCC |
| rck2∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 rck2::KanR | ATCC |
| sat4∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 sat4::KanR | ATCC |
| sip18∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 sip18::KanR | ATCC |
| sod1∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 sod1::KanR | ATCC |
| sod2∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 sod2::KanR | ATCC |
| ssk2∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 ssk2::KanR | ATCC |
| ssk22∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 ssk22::KanR | ATCC |
| tps1∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 tps1::KanR | ATCC |
| tps2∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 tps2::KanR | ATCC |
| tps3∆ | MATahis3∆1 leu2∆0 met15∆0 ura3∆0 tps3::KanR | ATCC |
| S. pastorianus | ||
| C. albicans (BC15) | Clinical isolate | Brendan Cormack |
| C. glabrata (BC615) | Clinical isolate | Brendan Cormack |
Media and growth conditions
Standard rich media were prepared as described previously (Rose et al. 1990). Assay buffer for desiccation tolerance assays was either deionized reverse-osmosis water or a ×8 dilution of phosphate buffered saline: NaCl, 17.1 mm; KCl, 0.338 mm; Na2HPO4, 1.25 mm; KH2PO4, 0.220 mm; pH 7.4.
Anaerobic culture was performed in an Invivo2 400 hypoxic workstation (Biotrace) at 23°. A test-tube rack secured to a vortexer was used to agitate 5-ml cultures. Reagents and equipment were deoxygenated by preconditioning for 24 hr in the workstation. To support anaerobic growth of S. cerevisiae, YEPD was supplemented with 0.5% Tween-80 (Cole-Parmer) and 20 μg/ml ergosterol (Sigma) (Andreasen and Stier 1953, 1954). Log-phase cultures were initiated from overnight cultures grown anaerobically. Saturated anaerobic cultures were obtained by 48 hr of anaerobic incubation. Desiccation was performed by anaerobic air drying in 96-well plates.
Quiescent and nonquiescent fractions of a 7-day culture were obtained as described previously (Allen et al. 2006), except that fractions were washed in dilute PBS. Generation of ρ− strains by treatment with ethidium bromide was performed as previously described (Goldring et al. 1971).
Quantitative assay
Cultures were incubated for 48 hr at 30°. Samples were prepared by washing ∼107 cells twice in 1 ml assay buffer and bringing the final volume to 1 ml. Undesiccated controls were plated for colony counting. For vacuum desiccation, two 100-µl aliquots were transferred to 1.5-ml microcentrifuge tubes and pelleted at 14,000 rpm in a microcentrifuge. A total of 85 µl of supernatant was removed, and the tubes were subjected to vacuum desiccation (Speedvac Concentrator SVC 100H, Savant Systems), without added heat, for between 5 and 24 hr. For air drying, two 100-µl aliquots were transferred to wells of a 96-well plate (Becton Dickinson, 353075), and allowed to desiccate in a 23° incubator, with the lid raised (Supporting Information, Figure S2), for 48 hr. Samples were resuspended in assay buffer and plated for colony counting. Data were entered into a spreadsheet (Microsoft Excel 2004 for Mac version 11.2), and the number of colony forming units per milliliter (CFU/ml) for each plate was computed. For each experiment, CFU/ml for the two controls was averaged. The relative viability of each of the two experimental samples was determined by dividing the CFU/ml for that sample by the average CFU/ml of the control plates. These two relative viability values were then averaged and their standard deviation was computed using the STDEVP worksheet function.
Semiquantitative assay
Samples were prepared and desiccated as described (see Materials and Methods, Quantitative assay). Five serial ×10 dilutions of each control or sample were prepared in 96-well plates, and 5 µl of each of the six dilutions were sequentially spotted to YEPD/agar.
Data normalization
To combine values obtained from more than one experiment, relative viability numbers for experimental samples are divided by the relative viability obtained from the wild-type internal control assayed in the same experiment, and are reported as “normalized percent relative viability.”
Comparative genomic hybridization
Genome-wide detection of single nucleotide polymorphisms was performed as described previously (Gresham et al. 2006).
Determination of complementation groups
Each of the 14 revertant MATa ρ− strains was crossed with each of the 12 revertant recessive MATα ρ− strains. The diploid progeny of these crosses were assayed semiquantitatively (see Materials and Methods, Semiquantitative assay) with vacuum desiccation. The parent strains were assigned to the same complementation group if the diploid was desiccation tolerant.
Genome-wide screen
Using a replicate of the MATα deletion collection in 96-well plate format, each plate was replicated with a pronged manifold into YEPD (75 µl/well). After 2 days of growth at 30°, each culture was replicated onto YEPD/agar as an undesiccated control. The lid of the plate was then elevated to allow the culture to air dry (Figure S4). The desiccated wells were rehydrated and replicated to YEPD/agar (Figure S3).
Respiration inhibition
Myxothiazol (Thierbach and Reichenbach 1981) was applied at a concentration of 30 μg/ml from a 1 mg/ml stock solution of the inhibitor in methanol. To phenocopy the ρ− mutation, a single colony of wild-type yeast was inoculated into 5 ml of treated YEPD and grown to saturation, and the assay buffer used to wash and desiccate the cells was also treated with inhibitor. To inhibit respiration during growth only, 1 ml of the inhibited culture was washed twice in 1 ml of inhibitor-free assay buffer before desiccation. To inhibit respiration during desiccation only, a sample prepared from an untreated saturated culture was washed and desiccated in assay buffer containing inhibitor.
Generation of spontaneous revertants
A total of 5 ml YEPD was inoculated from a single colony of ρ− yeast. Cultures were incubated for 48 hr, then desiccated under vacuum. A total of 100 µl of a 101× dilution of the desiccated sample was spread on YEPD agar and incubated at 30° for 48 hr. The colonies that developed were pooled by transferring 1 ml YEPD to the surface of the plate and suspending the colonies in that liquid. A total of 50 µl of this suspension was used to inoculate 5 ml of liquid YEPD and the growth/desiccation cycle was repeated. A total of 5 μg/ml ampicillin was added in further rounds to inhibit contaminaion. When hundreds or thousands of colonies developed on a plate after desiccation rather than the dozens usually observed, single colonies from that plate were cultured individually and assayed for desiccation tolerance.
Inhibition of translation and transcription
Samples were prepared as described (see Materials and Methods, Quantitative assay). Transcription: 1 µl each of DMSO and a 5 mg/ml stock solution of thiolutin (Pfizer) in DMSO were added to produce a final concentration of 5 μg/ml thiolutin and 0.2% DMSO. Translation: 2 µl of a 5 μg/ml stock solution of cycloheximide in DMSO were added to produce a final concentration of 10 μg/ml cycloheximide and 0.2% DMSO. Untreated controls were brought to 0.2% DMSO by adding 2 µl solvent to the sample. Treated and untreated samples were then assayed using vacuum desiccation.
Optical density
Measurements were taken on a Gilford Stasar III spectrophotometer at a wavelength of 600 nm. Dense cultures were diluted to achieve an optical density of between 0.100 and 0.500.
Results
Characterization of desiccation tolerance in wild-type budding yeast
To measure desiccation tolerance in S. cerevisiae, we developed a quantitative colony counting assay to determine the proportion of cells in a growing culture that survive desiccation. Briefly, cultures are harvested and brought to a cell density of between 0.5 × 108 and 1.0 × 108 cells/ml by either dilution or concentration, depending on the original cell density. An aliquot is removed to determine cell viability prior to desiccation. The remaining culture is desiccated, samples are rehydrated and then assayed for viability. The ratio of the viability of the desiccated sample to undesiccated control provides a measure of the desiccation tolerance and is expressed as percent relative viability.
The percent relative viability after desiccation is ∼5–20% for saturated cultures of typical laboratory wild-type yeast strains grown for 48 hr in YEPD. During development of this assay we noticed that there is no difference in relative viability obtained by desiccating samples rapidly under vacuum centrifugation or slow air drying (Figure S1). However, the percent relative viability can differ two- to five-fold for wild-type cultures in different desiccation experiments, potentially reflecting subtle differences in growth or desiccation conditions. Therefore, all desiccation experiments include a wild-type saturated culture as an internal standard. Normalization to the wild-type culture in each experiment allows comparison between cultures in different desiccation experiments. It should be noted that these normalizations are usually irrelevant as our conclusions are based upon differences in relative viability of many orders of magnitude between cultures. With this assay, we began our studies by characterizing how desiccation tolerance varies with cell growth, the time spent in the desiccated state, and speciation.
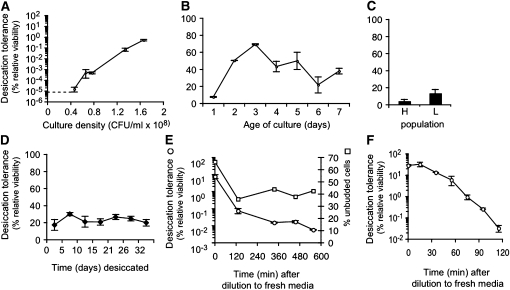
To address how desiccation tolerance varies as a function of cell growth, we established a wild-type culture in log phase (6.0 × 106 cells/ml), and quantitatively assayed it for desiccation tolerance at intervals until it reached late log phase (1.7 × 108 cells/ml) (Figure 1A). The percent relative viability is <10−5 at cell densities <4.6 × 107 cells/ml. Above this density, the percent relative viability rises with increased cell density, eventually reaching a value of 0.5%. These data establish a dramatic increase in desiccation tolerance at late log phase.
Figure 1 .
Culture of yeast consists of sensitive and tolerant populations, whose relative proportions are governed by growth phase. (A) Log-phase culture of BY4742 was established in YEPD at ∼106 cells/ml, then assayed for desiccation tolerance at various times as it grew to 1.8 × 108 cells/ml. Dashed line indicates lower limit of relative viability detectable by the assay as performed. Desiccation tolerance is expressed as the percent relative viability, the colony forming units of the desiccated and rehydrated culture divided by the undesiccated control (see Results). (B) Yeast culture was inoculated in YEPD then assayed for desiccation tolerance at 1-day intervals for 7 days. (C) Seven-day-old culture of BY4742 was separated into heavy (H) (quiescent) and light (L) populations (see Materials and Methods), which were then assayed for desiccation tolerance. (D) Several desiccated samples of yeast were prepared from a saturated culture and assayed at intervals of several days for ∼1 month. (E) Saturated yeast culture was inoculated into YEPD at 2 × 106 cells/ml and assayed for desiccation tolerance at various times for several hours. (F) Saturated yeast culture was inoculated into YEPD at 2 × 106 cells/ml and then assayed for desiccation tolerance at ∼15-min intervals for 2 hr.
Since desiccation tolerance increases from late log phase to saturation, reaching 20% after 48 hr, we wondered whether the proportion of tolerant cells would continue to increase in cultures grown for longer than 48 hr. We inoculated a culture of wild-type cells in YEPD and assayed it every 24 hr for 7 days (Figure 1B). Percent relative viability rises to a maximum of 69% at 3 days, then slowly declines to 20%. Since cells in a saturated culture have stopped dividing, this finding indicates that changes in metabolism within the cell can have a measurable effect on desiccation tolerance. Furthermore even after prolonged incubation, the saturated culture persists as a mixture of desiccation tolerant and sensitive cells.
Precedence exists for two populations of cells within saturated yeast cultures. Density sedimentation of saturated cultures reveals a light and heavy cell population; the latter has severely reduced metabolism and has been characterized as quiescent (Allen et al. 2006). We wondered whether the quiescent population might correspond to the desiccation tolerant cells. We separated a 7-day-old culture of wild-type yeast into light and heavy bands and assayed each band separately (Figure 1C). Cells in the heavy and light bands show equivalent desiccation tolerance. Thus desiccation tolerance appears to be independent of quiescence.
We next addressed how stable the desiccation tolerant state is in saturated cultures. First we asked whether desiccated cells lose viability with time after they are desiccated. We simultaneously desiccated multiple samples of a saturated culture of wild-type yeast, then rehydrated them at different intervals over 1 month and assayed them for viability (Figure 1D). The percent relative viability remains ∼20% independent of the time spent in the desiccated state. We conclude that the desiccated state is indeed stable and that desiccated cells are able to prevent or mitigate any further damage induced during storage under ambient conditions.
Second, we asked how stable are the desiccation tolerant cells in saturated cultures upon return to exponential growth. Saturated cultures are used as inocula for exponential cultures. Since the exponential cultures are very desiccation sensitive, the desiccation tolerant cells in the saturated inocula must either lose their tolerance or retain their tolerance and be diluted out by newly generated daughter cells and their progeny. To distinguish between these possibilities, we diluted a saturated culture to a density of 4.1 × 106 cells/ml in fresh media and then maintained the culture at this density for multiple divisions by rediluting the culture each time it doubled. We assayed desiccation tolerance of the culture immediately upon dilution from saturation and at regular intervals thereafter. Cells rapidly begin to lose their desiccation tolerance, dropping by two orders of magnitude within 2 hr, then slowly continuing to lose tolerance for several more hours, a biphasic or perhaps logarithmic response possibly related to the concentration of dextrose or ethanol in the medium. The proportion of unbudded cells declines from ∼70% to just <50%, indicating that cell division had begun (Figure 1E). To obtain a higher resolution measure of the kinetics of tolerance loss, we repeated the experiment, taking samples at more frequent intervals (Figure 1F). Tolerant cells begin to become sensitive within 30 min of being introduced at low density into YEPD. This rapid loss of desiccation tolerance when the culture has not yet even doubled demonstrates that the tolerant cells in the saturated inoculum must lose their tolerance upon exposure to their new growth conditions. Thus yeast cells can transit from the tolerant to the sensitive state as well as from the sensitive to the tolerant state (Figure 1A).
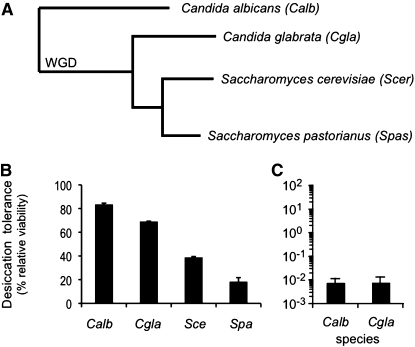
Saccharomycotina, the taxon of ascomycetes to which S. cerevisiae belongs, may be a desiccation-tolerant taxon, or S. cerevisiae may be unique in possessing this trait. We therefore grew saturated 48-hr cultures of several members of Saccharomycotina (Figure 2A) and assayed them for desiccation tolerance (Figure 2B). The tested members of the clade are all desiccation tolerant (with between 20 and 80% relative viability). The level of desiccation tolerance does not appear to correlate with phylogenetic distance from S. cerevisiae or relationship to the whole genome duplication. To further investigate the evolutionary relationship between desiccation tolerance and phylogenetic distance from S. cerevisiae, log-phase cultures of Caenorhabditis albicans and C. glabrata were established and also found to exhibit sensitivity, being four orders of magnitude more sensitive than when saturated (Figure 2C). We conclude that Saccharomycotina is likely a highly desiccation tolerant taxon, with tolerance and sensitivity governed by a similar set of effectors.
Figure 2 .
Desiccation tolerance is conserved among the phylogenetic neighbors of S. cerevisiae. (A) Partial phylogenetic tree of Saccharomycotina. “WGD” indicates the branch evolving after the whole genome duplication. (B) Saturated cultures of Candida albicans, C. glabrata, Saccharomyces cerevisiae, and S. pastorianus were assayed for desiccation tolerance. (C) Log-phase cultures of C. albicans and C. glabrata were assayed for desiccation tolerance.
Testing the predicted effectors and stress pathways for their importance in desiccation tolerance
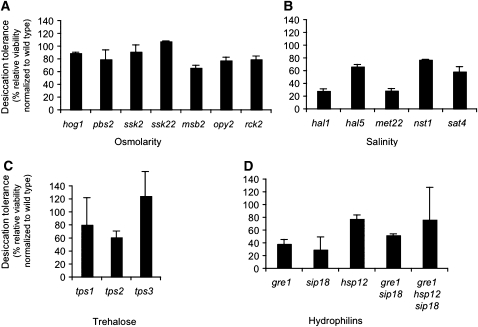
Loss of water increases the solute to solvent ratio in a cell, thereby changing osmolarity and salt concentration. Yeast cells maintain homeostasis in salinity and osmolarity through regulators that induce or change the activity of membrane pumps. They also produce small molecules such as trehalose and hydrophilins that may act as osmolytes and/or water substitutes (Yancey et al. 1982; Oliver et al. 2001; Crowe 2002). To test whether any of these known functions are important for desiccation tolerance we measured desiccation tolerance in deletion mutants that eliminate their function (Figure 3). Desiccation tolerance is either unaffected or reduced only a few fold in deletions of genes encoding master regulators or effectors of osmolarity (Figure 3A), salinity (Figure 3B), trehalose biosynthesis (Figure 3C), and three hydrophilins (Figure 3D). These three hydrophilins were selected because a previous study suggested they were highly induced during desiccation. Even a triple mutant lacking all three hydrophilins exhibited normal desiccation tolerance. None of these deletion strains reduce desiccation tolerance anywhere near the five orders of magnitude reduction seen for exponential growth (Figure 1A). Therefore, either changes in osmolarity and/or salinity are not major causes of desiccation sensitivity, or yeast cells have alternative yet-to-be-discovered pathways redundant with these canonical pathways that mitigate these stresses during desiccation.
Figure 3 .
Deletion of genes predicted to be important for desiccation tolerance have only a minimal impact on the trait. Saturated cultures of strains with deletions of genes involved in response to (A) osmolarity and (B) salinity were assayed for desiccation tolerance. (C) Saturated cultures of strains with deletions of genes required for trehalose synthesis were assayed for desiccation tolerance. (D) Saturated cultures of strains with single and multiple deletions of hydrophilins reported to be highly induced by desiccation were assayed for desiccation tolerance. All mutants listed in this figure and the remaining figures harbor deletion alleles constructed by the whole genome yeast consortium (Winzeler et al. 1999).
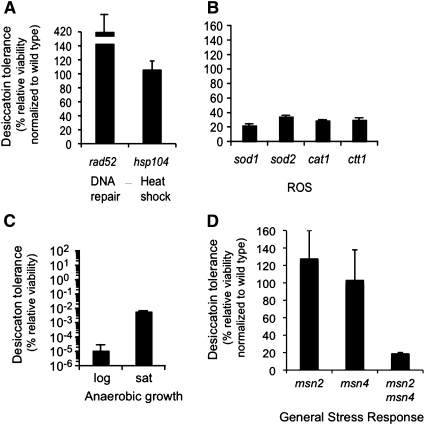
Desiccation has also been proposed to stress cells by causing DNA double strand breaks (DSB), protein denaturation, or ROS. Double strand breaks are repaired by a number of pathways all of which require Rad52p. Protein aggregates are resolubilized primarily by Hsp104p (Lee et al. 2010). However, deletion of neither RAD52 nor HSP104 reduces desiccation tolerance (Figure 4A). Oxidative stress is mitigated in yeast primarily by two superoxide dismutases and two catalases. Individual knockouts of each of these genes generated a fivefold reduction in viability (Figure 4B). Since multiple knockouts of these ROS scavengers tend to produce very sick strains, this line of exploration was not pursued via a multiple deletion strategy. However, if ROS are the major cause of desiccation sensitivity, we should be able to generate tolerance by propagating yeast anaerobically where the formation of ROS is abrogated (Figure 4C). The absence of oxygen does not rescue the extreme sensitivity of log-phase cells and in fact induced three orders of magnitude increase in sensitivity in saturated cultures. From these findings we conclude that although reduction of ROS scavenging ability does introduce a mild phenotype, molecular oxygen is in fact required to generate significant desiccation tolerance in saturated cultures of yeast.
Figure 4 .
Ablation of stress responses predicted to contribute to desiccation tolerance minimally impact the trait, but anaerobic growth induces sensitivity. Saturated cultures of strains with deletions of genes responsible for DSB repair (A), heat shock tolerance (A), mitigation of ROS stress (B), and induction of the general stress response (D) were assayed for desiccation tolerance (C). Anaerobically cultured log-phase and saturated cultures of wild-type yeast were assayed for desiccation tolerance (see Materials and Methods).
Since inactivation of specific stress response pathways has at most only a minor impact on tolerance, we asked whether tolerance would be compromised by the simultaneous downregulation of multiple stress response pathways. Many of these stress pathways are regulated by the Msn2p and Msn4p paralogs, the primary transcriptional regulators of the general stress response in yeast. While the msn2∆ and msn4∆ strains exhibit no defect in desiccation tolerance, tolerance is reduced about fivefold in the msn2∆msn4∆ double mutant (Figure 4D). Thus at least one factor regulated by the established stress responses contributes to desiccation tolerance. In summary, most of desiccation tolerance must be mediated by factors either redundant with or independent of canonical stress responses previously correlated with desiccation.
Genome-wide screen for sensitive mutants identifies respiration as a prerequisite for desiccation tolerance
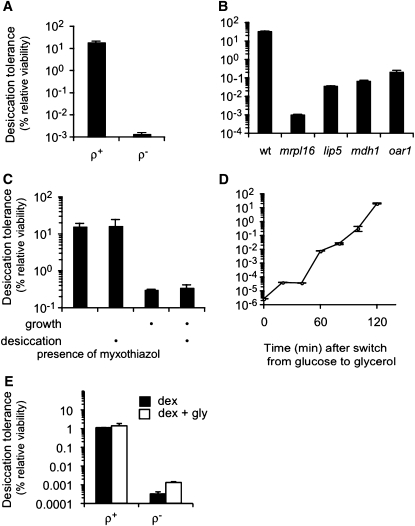
Since testing candidates for effectors of desiccation tolerance failed to identify genes whose deletion produces sensitivity approaching that of log-phase cultures, an unbiased genetic screen of mutants for desiccation sensitivity might reveal pathways that had heretofore been unappreciated as contributors to desiccation tolerance. We screened the haploid deletion collection of budding yeast for desiccation sensitivity (Materials and Methods, Genome-wide screen). We originally found ∼100 deletion strains that appeared promising, but on further characterization we determined that most of these were ρ− respiration-incompetent strains. Indeed, all ρ− strains tested (except the spontaneous revertants described below) were found to be desiccation sensitive. The mutant mrpl16∆ is ρ− and is used as a representative ρ− strain. In addition, we generated ρ− wild type by treatment with ethidium bromide. The ρ− wild-type cells exhibit the same dramatic decrease in desiccation tolerance (Figure 5A). Thus mitochondrial function is required for desiccation tolerance.
Figure 5 .
Respiration is required for acquisition of desiccation tolerance. An unbiased screen of the haploid deletion collection was performed to identify mutants sensitive to desiccation when grown to saturation. (A) ρ+ and ρ– BY4742 were grown to saturation and assayed for desiccation tolerance (see Materials and Methods). (B) Saturated cultures of mutants recovered from the haploid deletion collection with at least two orders of magnitude reduction in desiccation tolerance. (C) Saturated cultures of BY4742 were exposed to 0.2 µg/ml myxothiazol during growth, during desiccation, or during both growth and desiccation (as indicated by the bullet) and then assayed for desiccation tolerance. (D) Glucose fermenting log-phase culture of BY4742 was forced to respire by switching the growth medium to glycerol, a nonfermentable carbon source. Samples were taken approximately every 20 min and assayed for desiccation tolerance. (E) ρ+ and ρ– BY4742 cultures were grown to saturation and assayed for desiccation tolerance either in YEPD (solid bars) or in YEPD with 3% glycerol added (open bars).
To eliminate ρ− mutants as a cause of desiccation sensitivity, we isolated respiratory competent cells for each candidate deletion by demanding their growth on glycerol. Of these, only three strains were found to generate sensitivity approaching that of log-phase cells: lip5∆, mdh1∆, and oar1∆ (Figure 5B). Introduction of lip5∆, mdh1∆, and oar1∆ deletions into the A364A background also results in a desiccation-sensitive phenotype (Figure S5), indicating that desiccation tolerance requires these genes. All three of these genes have a connection with respiration or mitochondria: LIP5 codes for lipoic acid synthase; lipoic acid is an essential coenzyme of pyruvate dehydrogenase, which contributes to transforming pyruvate into acetyl CoA. MDH1 codes for the mitochondrial malate dehydrogenase, which performs the penultimate step of the tricarboxylic acid cycle. OAR1 codes for a enzyme involved in mitochondrial fatty acid metabolism. All three deletion strains, while capable of growth on glycerol, do so very slowly. Taken together, our results suggest that respiration is required for saturated cultures to acquire desiccation tolerance. This conclusion explains the requirement for oxygen that we observed previously (Figure 4C) and the transition from sensitivity to tolerance in late exponential cultures (Figure 1A), which correlates with the diauxic shift when cells switch from fermentation to respiration.
To further explore the connection between respiration and desiccation tolerance, we asked when the ability to respire is required for tolerance: before, during, or both before and during desiccation. For this purpose, we applied the respiration inhibitor myxothiazol (Materials and Methods, Respiration inhibition) either during growth only, during desiccation only, or for the duration of the entire experiment. As expected, inhibition of respiration by myxothiazol during both growth and desiccation phenocopies the desiccation sensitivity of ρ− cells. A similar level of desiccation sensitivity is observed when respiration is inhibited only during growth. However normal desiccation tolerance is observed when respiration is inhibited only during desiccation (Figure 5C). Thus to acquire desiccation tolerance, cells must have experienced respiration prior to desiccation but need not respire during desiccation itself.
To determine how rapidly yeast cells can acquire desiccation tolerance by respiring, we exploited the fact that our log-phase cultures are very desiccation sensitive when grown on a medium with glucose, a fermentable carbon source (Figure 1A). We replaced this medium with a medium containing glycerol, a carbon source that can be assimilated only via respiration. Within 2 hr (within one doubling), the dividing cultures increase in desiccation tolerance by seven orders of magnitude (Figure 5D). To test that this acquisition of desiccation tolerance is through the respiration of glycerol rather than some other effect of glycerol, we added glycerol directly to the medium containing glucose (Figure 5E). The presence of glycerol in the medium does not rescue the desiccation tolerance of the ρ− cells, indicating that glycerol only induces tolerance when it is used as a carbon source. In summary, the cellular changes required to acquire desiccation tolerance can occur within one cell division and dividing cells can exhibit the same level of tolerance as stationary cells (compare Figures 5D and 1A).
Transcriptional regulation of desiccation tolerance
Some molecular component of respiration may be required directly for desiccation tolerance or respiration may provide a signal that activates a cascade leading to tolerance. If the latter is true, then it should be possible to isolate mutations that activate desiccation tolerance via bypass of the respiration signal. Therefore, starting with 14 independently established cultures of ρ− MATa (BY4741: mrpl16∆; ρ−) and ρ− MATα (BY4742: mrpl16∆; ρ−) strains, we grew each culture to saturation, desiccated them, and then pooled the colonies that survived desiccation and repeated the process. After four rounds of this procedure, strains with almost wild-type desiccation tolerance were generated. Of the 28 strains recovered, two of the MATα strains were found to be dominant mutations while the remaining strains were recessive. The existence of these revertants indicates that a direct product of respiration is not essential for desiccation tolerance. None of the revertants were competent for growth on glycerol, indicating that the mutations did not restore respiratory competence.
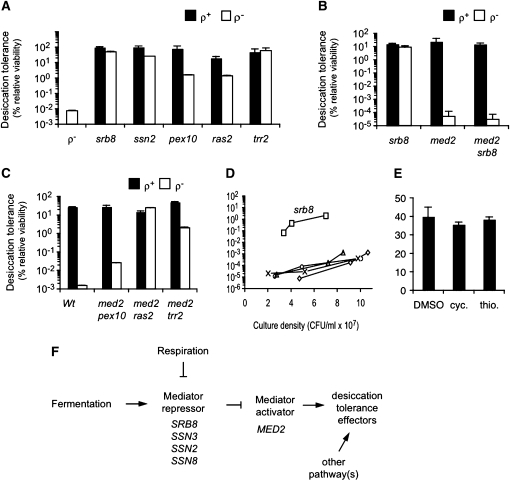
To identify the mutations responsible for rescuing the desiccation sensitivity of ρ− yeast, we assigned 21 of the recessive revertants to five complementation groups (Table 2). We then hybridized genomic DNA from each strain to a tiling microarray, a technique capable of identifying loci that are polymorphic relative to the reference genome sequence. To eliminate irrelevant or false positive polymorphisms, we focused on polymorphisms that co-occurred within a single gene for each complementation group. We sequenced these polymorphic alleles and in almost all cases they generated a nonsense codon in the gene. For these candidates, we constructed ρ− variants of the corresponding deletion strain from the yeast deletion library and assayed them for desiccation tolerance. The ρ− variants are desiccation tolerant, providing further evidence that the inactivation of each of these genes is responsible for the rescue of the desiccation sensitivity of ρ− strains (Figure 6A).
Table 2 . Complementation group assignments of recessive spontaneous revertants.
| Locus | Number of mutants | Function |
|---|---|---|
| SRB8 | 5 | Mediator repressive module |
| SSN2 | 4 | Mediator repressive module |
| PEX10 | 1 | Peroxisome import |
| RAS2 | 5 | Small GTP binding protein |
| TRR2 | 6 | Mitochondrial thioredoxin reductase |
| Unknown | 5 |
Figure 6 .
Desiccation tolerance arises from at least two pathways, at least one of which is transcriptionally regulated. Desiccation tolerant revertants of ρ− BY4742 were isolated, and whole genome sequencing of the revertants revealed putative null alleles responsible for the restoration of desiccation tolerance (see Materials and Methods and Results). (A) Deletion alleles from the haploid deletion collection that correspond to the revertant alleles were identified. Saturated cultures of the ρ+ and ρ− variants of these deletions allele were assayed for desiccation tolerance. (B and C) Saturated cultures of the ρ+ and ρ− variants of mutants in the Mediator repressor (srb8) and activator (med2) submodules were assayed for desiccation tolerance. (D) Log-phase cultures of ρ+ BY4742 (diamond), pex10 (circle), ras2 (X’s), srb8 (square), trr2 (triangle) were assayed for desiccation tolerance. (E) Saturated cultures of BY4742 were desiccated in the presence of the translation inhibitor cycloheximide (cyc) or the transcription inhibitor thiolutin (thio) and then assayed for desiccation tolerance. (F) Summary of the regulation of desiccation tolerance in fermenting and respiring yeast.
Two of the deletions, srb8∆ and ssn2∆, identify subunits of the Mediator repressive module. Mediator is a large multisubunit complex that regulates transcription of hundreds of genes (Van De Peppel et al. 2005). The repressive module contains four subunits that together repress Mediator’s other subunits required for transcription activation. We assayed ρ− strains deleted for the other two components (SSN3 and SSN8) of the Mediator repressive module and found them to be desiccation tolerant as well (Figure S4). Thus loss of any subunit of the Mediator repressive module leads to restoration of desiccation tolerance in cells defective for respiration. This result suggests that factors important for desiccation tolerance in budding yeast are under transcriptional control.
The repressive module inactivates the activator module of Mediator that includes Med2p (Van De Peppel et al. 2005). We hypothesized that inactivation of the repressive module allows the activator module to induce genes necessary for desiccation tolerance. If so, inactivation of the activator module by deletion of MED2 in the srb8∆ background should eliminate the ability of srb8∆ to rescue the desiccation sensitivity of a ρ− strain. Indeed this is the case (Figure 6B). To determine whether the non-Mediator revertants, pex10∆, ras2∆, and trr2∆ operate in the same pathway, we assayed double knockouts of these genes with med2∆ and found that only the med2∆pex10∆ double mutant loses significant desiccation tolerance like the srb8∆med2∆ double mutant (Figure 6C). Thus MED2-independent as well as MED2-dependent pathways control desiccation tolerance. In agreement with this, saturated cultures of ρ+ med2∆ strains are not desiccation sensitive, indicating that desiccation tolerance can also be induced by MED2-independent mechanism(s).
Since the revertant deletions rescue sensitivity generated by damage to the mitochondrial genome in ρ− strains, we were curious whether they also rescued sensitivity in log phase when ρ+ strains are also extremely sensitive. We therefore established log-phase cultures of ρ+ strains of each of the deletants and assayed them for desiccation tolerance at cell densities for which wild-type strains are extremely sensitive (Figure 6D). The srb8∆ strain is extremely desiccation tolerant in these conditions, whereas none of the other deletions rescues sensitivity as they do in saturated ρ− strains. We conclude that disruption of the Mediator repressive module allows the transcription of genes whose products are instrumental in generating desiccation tolerance.
Similar to our earlier studies on respiration, we asked whether the transcriptional induction by Mediator of desiccation tolerant factors occurs at the time of desiccation or prior to desiccation. To distinguish between these two possibilities, we desiccated a saturated yeast culture in the presence of transcription or translation inhibitors (see Materials and Methods, Translation and transcription inhibition). In both cases, desiccation tolerance was unaffected (Figure 6E), supporting the conclusion that all the genes needed for desiccation tolerance are already transcribed and translated prior to desiccation. This finding is consistent with the concept that Mediator activation module activates expression of desiccation-relevant genes when respiration begins during the diauxic shift.
Discussion
Here we characterize the effect of growth conditions on desiccation tolerance in budding yeast. The fraction of cells surviving desiccation ranges from 20–70% in saturated cultures to fewer than one in a million in early log-phase cultures. This dramatic difference is observed in many different backgrounds, related species, and under both slow and rapid desiccation regimens. Previous studies have found that log phase are only 20-fold more sensitive than saturated cultures (Beker and Rapoport 1987; Ratnakumar and Tunnacliffe 2006) (Ratnakumar et al. 2011). Why these studies did not observe the six-orders-of-magnitude difference reported here is unclear, but the magnitude of the difference is critical, shaping our interpretation of results in this study and previous studies. It also makes the study of desiccation tolerance in yeast amenable to powerful genetic screens and selections as exemplified in this study.
With this new metric, we assessed candidate effectors of desiccation tolerance implicated by prior studies in yeast and other organisms. Remarkably, mutants lacking these proteins exhibit, at most, only small decreases in desiccation tolerance. We corroborate previous studies showing that neither trehalose biosynthesis (Ratnakumar and Tunnacliffe 2006) nor osmoregulation by Hog1p (Ratnakumar et al. 2011) are essential for robust desiccation tolerance. Additionally, we show that desiccation tolerance does not require DNA double strand break repair, the hydrophilins most highly expressed during desiccation (Singh et al. 2005), the major protein-folding chaperone Hsp104p, or the complete complement of ROS scavengers. The lack of sensitivity in these mutants suggests that cells might possess undiscovered pathways that operate in the absence of these canonical stress responses. Alternatively, these stresses may not be the lethal ones imposed by desiccation. Indeed, contrary to the prevailing consensus, we show that eliminating oxygen and thus ROS stress causes desiccation sensitivity rather than tolerance. If these stresses are not the lethal ones, then their potentially relevant stress response factors may serve functions other than protection from desiccation. For example recent studies suggest that trehalose might accumulate in stationary cells as a preferred carbon source to allow rapid exit from G0 when nutrients return (Shi et al. 2010).
Complementing our candidate testing of known stress responses, we performed an unbiased genome-wide screen for mutants that are extremely sensitive in saturated cultures, producing two remarkable findings. First, defects in most cellular processes do not dramatically sensitize yeast to desiccation. Some minor sensitivity may occur, as other recent studies have identified from ∼300 (Shima et al. 2008) to >500 (D’Elia et al. 2005) different deletion mutants in a number of cellular processes that produce small but measurable defects in tolerance. However, such small effects relative to fermenting cells could also be explained by indirect effects on fermentation or respiration. Desiccation tolerance is refractory to many cellular perturbations despite the reliance of nearly every biochemical process upon the presence of water.
Second, in contrast to the general robustness of desiccation tolerance, mutants in only one function, respiration, reduce desiccation tolerance by several orders of magnitude. The connection between respiration and tolerance is strongly supported by additional observations also reported here. A saturated culture of wild-type cells fails to exhibit desiccation tolerance when it is forced to grow to saturation without respiring, by either the removal of oxygen or the presence of respiratory inhibitors. Furthermore, the majority of wild-type cells acquire desiccation tolerance at a cell density when cells in the culture transit from fermentation to respiration (this study and Ratnakumar and Tunnacliffe 2006). Finally, early log-phase cultures can exhibit the same level of tolerance as saturated cultures if the log-phase cultures are forced to respire by growth on glycerol. Thus we provide compelling evidence that respiration triggers budding yeast to undergo a dramatic shift from desiccation sensitivity to tolerance.
While respiration is an important trigger for desiccation tolerance, a significant fraction of respiring cells remains sensitive (this study). Perhaps our laboratory conditions fail to mimic natural conditions that are more effective at inducing tolerance. Alternatively, the tolerant state may have a cost, with incomplete conversion of an entire culture to tolerance programmed to ensure maximization of survival options with regard to unanticipated changes to its environment. Additionally, desiccation tolerant cells continue to accumulate in cultures after reaching saturation and switching to respiration. These observations are consistent with processes other than respiration contributing to the acquisition of tolerance. Our study argues that quiescence is not one of these other processes. However, a recent study suggests that autophagy is required for desiccation tolerance, as mutants exhibit a 10-fold defect in tolerance (Ratnakumar et al. 2011). The induction of autophagy in saturated cultures and a similar fold increase in desiccation tolerance after saturation is consistent with a role for autophagy in tolerance.
Two observations support the notion that respiration triggers the acquisition of desiccation tolerance rather than directly providing a protective reagent or critical energy source. First, respiration inhibitors reduce tolerance when present during the growth of the culture but not when present only during desiccation. Second, we isolate mutants that achieve robust desiccation tolerance without respiring. Several possibilities potentially explain how respiration triggers desiccation tolerance. The stresses associated with respiration may mimic the stress conditions of desiccation. Respiration, a known prerequisite for completion of meiosis and sporulation, may be coupled to desiccation tolerance to ensure tolerance of spores. This coupling may be fortuitously preserved during vegetative growth. Alternatively, desiccation tolerance may be a constitutive state of yeast in the wild, because of the potential for rapid changes in environment (for example, direct sunlight rapidly dehydrating the surface occupied by a yeast colony). The cell only suppresses desiccation tolerance and other protective shields under favorable fermenting growth conditions, when the growth rate is maximized. Clearly, discriminating between these and other possibilities will require better understanding of the regulatory pathway and effectors of desiccation tolerance controlled by respiration.
We therefore isolated suppressors that restore desiccation tolerance to a respiration defective mutant. The majority of our suppressors are loss-of-function mutations, which are highly biased to inactivate negative regulators of desiccation effectors. Indeed, our analysis revealed that one such negative regulator is the inhibitory submodule of the Mediator complex, an important component of the RNA polymerase II holoenzyme (Davis et al. 2002). Furthermore, we show that this submodule likely acts by repressing transcription activation by the Med2p activator submodule. Thus we provide compelling evidence that a transcriptional program regulates the transition from desiccation sensitivity to tolerance in budding yeast (Figure 6F). Intriguingly, while the Med2p submodule activates hundreds of genes, it has been implicated in specifically regulating other stress responses including the response to iron deficiency and the general stress response pathway controlled by Msn2p/Msn4p (Van De Peppel et al. 2005). Transcriptional regulation of water homeostasis has precedent as drought tolerance in plants is also transcriptionally regulated (Moore et al. 2009; Wang et al. 2009). Finally, Med2p cannot be the only positive regulator of desiccation effectors, since desiccation tolerance can occur independent of Med2p function in saturated cultures of both wild-type and ras2∆ ρ− cells (this study). Thus the regulation of desiccation tolerance is complex.
Observations from this study provide a foundation to identify effectors of desiccation tolerance. First we show that desiccation sensitivity in respiratory defective cells is rescued by inactivation of TRR2 and PEX1. TRR2 and PEX10 encode proteins important for mitochondrial thioredoxin function and peroxisome biogenesis, respectively. Thioredoxin and perioxisomes both impact cellular redox, suggesting that effectors of redox homestasis may be important for desiccation tolerance. Furthermore, the knowledge that desiccation tolerance is transcriptionally regulated by Med2p (and likely other transcription programs) is also very exciting. Budding yeast has a long track record of successfully elucidating targets of transcriptional programs, particularly when coupled with powerful genetic screens and selections like those demonstrated here. Thus exploiting this transcriptional control and the connection to redox homeostasis provides important new avenues to identify the elusive effectors of desiccation tolerance.
Acknowledgments
The authors thank Peter Espenshade for use of his anaerobic workstation, Kyle Cunningham for the kind gift of myxothiazol, and Brendan Cormack for the kind gift of Candida strains. We thank Hugo Tapia and Aaron Welch for critical reading of the manuscript. Funding for this work came from Howard Hughes Medical Institute to D.E.K. and National Institutes of Health grant P50 GM071508 to M.D.
Literature Cited
- Al-Rubeai M., Fussenegger M., Sharfstein S. T., Shen D., Kiehl T. R., et al. , 2007. Molecular response to osmotic shock, pp. 213–236 Systems Biology. Springer-Verlag, The Netherlands [Google Scholar]
- Allen C., Buttner S., Aragon A. D., Thomas J. A., Meirelles O., et al. , 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 174: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almoguera C., Jordano J., 1992. Developmental and environmental concurrent expression of sunflower dry-seed-stored low-molecular-weight heat-shock protein and Lea mRNAs. Plant Mol. Biol. 19: 781–792 [DOI] [PubMed] [Google Scholar]
- Andreasen A. A., Stier T. J. B., 1953. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for growth in a defined medium. J. Cell. Comp. Physiol. 41: 23–36 [DOI] [PubMed] [Google Scholar]
- Andreasen A. A., Stier T. J. B., 1954. Anaerobic nutrition of Saccharomyces cerevisiae. II. Unsaturated fatty and requirement for growth in a defined medium. J. Cell. Comp. Physiol. 43: 271–281 [DOI] [PubMed] [Google Scholar]
- Bartels D., 2005. Desiccation tolerance studied in the resurrection plant craterostigma plantagineum1. Integr. Comp. Biol. 45: 696. [DOI] [PubMed] [Google Scholar]
- Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A. A., 2008. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148: 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beker M., Rapoport A., 1987. Conservation of yeasts by dehydration, pp. 127–171 Biotechnology Methods. Springer-Verlag, Berlin [Google Scholar]
- Crowe J. H., 1972. Evaporative water loss by tardigrades under controlled relative humidities. Biol. Bull. 142: 407–416 [Google Scholar]
- Crowe J. H., Hoekstra F. A., Crowe L. M., 1992. Anhydrobiosis. Annu. Rev. Physiol. 54: 579–599 [DOI] [PubMed] [Google Scholar]
- Crowe L. M., 2002. Lessons from nature: the role of sugars in anhydrobiosis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 131: 505–513 [DOI] [PubMed] [Google Scholar]
- D’Elia R., Allen P. L., Johanson K., Nickerson C. A., Hammond T. G., 2005. Homozygous diploid deletion strains of Saccharomyces cerevisiae that determine lag phase and dehydration tolerance. Appl. Microbiol. Biotechnol. 67: 816–826 [DOI] [PubMed] [Google Scholar]
- Davis J. A., Takagi Y., Kornberg R. D., Asturias F. J., 2002. Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol. Cell 10: 409–415 [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C., 2008. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Gadd G. M., Chalmers K., Reed R. H., 1987. The role of trehalose in dehydration resistance of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 48: 249–254 [Google Scholar]
- Garay-Arroyo A., Colmenero-Flores J. M., Garciarrubio A., Covarrubias A. A., 2000. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 275: 5668–5674 [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., et al. , 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring E. S., Grossman L. I., Marmur J., 1971. Petite mutation in yeast. J. Bacteriol. 107: 377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal K., Walton L. J., Browne J. A., Burnell A. M., Tunnacliffe A., 2005. Molecular anhydrobiology: identifying molecules implicated in invertebrate anhydrobiosis. Integr. Comp. Biol. 45: 702. [DOI] [PubMed] [Google Scholar]
- Gresham D., Ruderfer D. M., Pratt S. C., Schacherer J., Dunham M. J., et al. , 2006. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science 311: 1932–1936 [DOI] [PubMed] [Google Scholar]
- Jiang M., Zhang J., 2002. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 53: 2401–2410 [DOI] [PubMed] [Google Scholar]
- Kranner I., Birtić S., 2005. A modulating role for antioxidants in desiccation tolerance. Integr. Comp. Biol. 45: 734–740 [DOI] [PubMed] [Google Scholar]
- Lee S., Sielaff B., Lee J., Tsai F. T. F., 2010. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc. Natl. Acad. Sci. USA 107: 8135–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesaffre Yeast Corporation, 2010. “Red Star Original” (http://www.redstaryeast.com/8/24/2010)
- Moore J. P., Le N. T., Brandt W. F., Driouich A., Farrant J. M., 2009. Towards a systems-based understanding of plant desiccation tolerance. Trends Plant Sci. 14: 110–117 [DOI] [PubMed] [Google Scholar]
- O’Rourke S. M., Herskowitz I., O’Shea E. K., 2002. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 18: 405–412 [DOI] [PubMed] [Google Scholar]
- Oliver A. E., Leprince O., Wolkers W. F., Hincha D. K., Heyer A. G., et al. , 2001. Non-disaccharide-based mechanisms of protection during drying. Cryobiology 43: 151–167 [DOI] [PubMed] [Google Scholar]
- Ratnakumar S., Tunnacliffe A., 2006. Intracellular trehalose is neither necessary nor sufficient for desiccation tolerance in yeast. FEMS Yeast Res. 6: 902–913 [DOI] [PubMed] [Google Scholar]
- Ratnakumar S., Hesketh A., Gkargkas K., Wilson M., Rash B. M., et al. , 2011. Phenomic and transcriptomic analyses reveal that autophagy plays a major role in desiccation tolerance in Saccharomyces cerevisiae. Mol. Biosyst. 7: 139–149 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F. M., Heiter P., 1990. Methods in Yeast Genetics: a Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sales K., Brandt W., Rumbak E., Lindsey G., 2000. The LEA-like protein HSP 12 in Saccharomyces cerevisiae has a plasma membrane location and protects membranes against desiccation and ethanol-induced stress. Biochimica et Biophysica Acta (BBA) -. Biomembranes 1463: 267. [DOI] [PubMed] [Google Scholar]
- Shi L., Sutter B. M., Ye X., Tu B. P., 2010. Trehalose is a key determinant of the quiescent metabolic state that fuels cell cycle progression upon return to growth. Mol. Biol. Cell 21: 1982–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima J., Ando A., Takagi H., 2008. Possible roles of vacuolar H+-ATPase and mitochondrial function in tolerance to air-drying stress revealed by genome-wide screening of Saccharomyces cerevisiae deletion strains. Yeast 25: 179–190 [DOI] [PubMed] [Google Scholar]
- Singh J., Kumar D., Ramakrishnan N., Singhal V., Jervis J., et al. , 2005. Transcriptional response of Saccharomyces cerevisiae to desiccation and rehydration. Appl. Environ. Microbiol. 71: 8752–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sømme L., 1996. Anhydrobiosis and cold tolerance in tardigrades. Eur. J. Entomol. 93: 349–357 [Google Scholar]
- Thierbach G., Reichenbach H., 1981. Myxothiazol, a new antibiotic interfering with respiration. Antimicrob. Agents Chemother. 19: 504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe A., Ricci C., 2006. Mechanisms of desiccation tolerance, pp. ESF LESC Exploratory Workshop, edited by Tunnacliffe A. European Science Foundation, Pembroke College, Cambridge, UK [Google Scholar]
- van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T. T. J. P., van Leenen D., et al. , 2005. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19: 511. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhu Y., Wang L., Liu X., Liu Y., et al. , 2009. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 230: 1155–1166 [DOI] [PubMed] [Google Scholar]
- Wehmeyer N., Hernandez L. D., Finkelstein R. R., Vierling E., 1996. Synthesis of small heat-shock proteins is part of the developmental program of late seed maturation. Plant Physiol. 112: 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Wright J. C., 1988. Structural correletes of permeability and tun formation in tardigrade cuticle: an image analysis study. J. Ultrastruct. Mol. Struct. Res. 101: 23–39 [Google Scholar]
- Yancey P. H., Clark M. E., Hand S. C., Bowlus R. D., Somero G. N., 1982. Living with water stress: evolution of osmolyte systems. Science 217: 1214–1222 [DOI] [PubMed] [Google Scholar]
- Zhu J.-K., 2002. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]