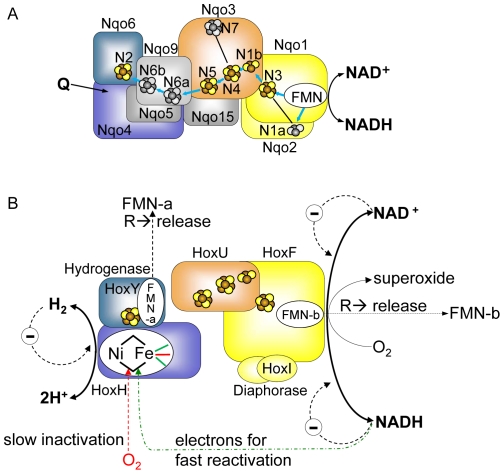
Figure 1. Modular structure and proposed cofactor arrangement and function of the soluble NAD+-reducing [NiFe]-hydrogenase of R. eutropha based on the results of the present study and references [7] , [15], [16], [21], [51].
Panel A displays the cofactors of the hydrophilic part of T. thermophilus Complex I [14]. Orthologous subunits of Complex I and SH carry the same colours. Iron-sulfur clusters, which are conserved in the SH, are colored in yellow/brown, those which are not present in the SH are shown in grey. The proposed electron transfer chain in Complex I is indicated by blue arrows. The N2 cluster localized in the Nqo6 subunit of Complex I corresponds to the iron-sulfur cluster in the hydrogenase small subunit HoxY (see Figs. S1,S2,S3). The proposed quinone-binding site (Q), which is situated in Nqo4, is indicated by an arrow [17]. Panel B shows the current SH model, the proposed localization of the individual cofactors and reactions taking place at the SH. The CN− ligands and the CO ligand of the Ni-Fe active site iron are shown in green and red, respectively. For sake of clarity, the hydrogenase and diaphorase modules are drawn separately. Main physiological reactions are shown in bold lines and fonts. “R” stands for “under reducing conditions”, “Θ” for “product inhibition”. Dashed lines represent effects, but without information of effect location. NADH-derived electrons for fast reactivation of the oxidized active site are passed through the FMN cofactors and the FeS clusters. For details see text and references [7], [15], [16], [21], [51].