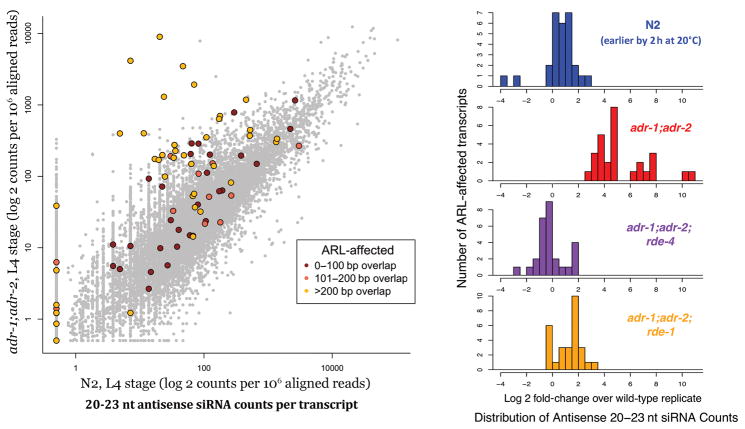
Figure 6. Accumulation of a second population of siRNAs in cis to ARLs.
(a) Genome-wide characterization of antisense siRNAs isolated using a 5′ phosphate independent capture protocol. Secondary siRNAs (antisense, 20–23nt) were tallied over each transcript. Transcripts overlapping ARLs are colored in dark red, orange, and yellow, based on the size of the total overlap (one, two, or three or more 100 bp segments, respectively). Genes that overlapped ARLs by at least three 100 bp segments and that exhibited a minimum 6-fold increase of secondary siRNAs in adr-1;adr-2 samples were deemed “ARL-affected transcripts”, and were considered as potential beneficiaries of ADAR-RNAi competition (without ADAR, they would be subject to populations of siRNAs produced by the RNAi machinery). (b) Effects of RNAi mutants on the secondary siRNA levels of ARL-affected transcripts. Antisense siRNA counts for ARL-affected transcripts were normalized to their respective wild-type levels. The top row (blue) denotes a separate biological preparation of wild-type animals at an earlier L4 stage (2 h earlier at 20°C). Secondary siRNA levels return to wild-type levels in adr-1;adr-2;rde-4 and adr-1;adr-2;rde-1 triple mutants. Gene-by-gene comparisons of expression-changes between different mutant backgrounds are depicted in Supplementary Figure 5.