Figure 1.
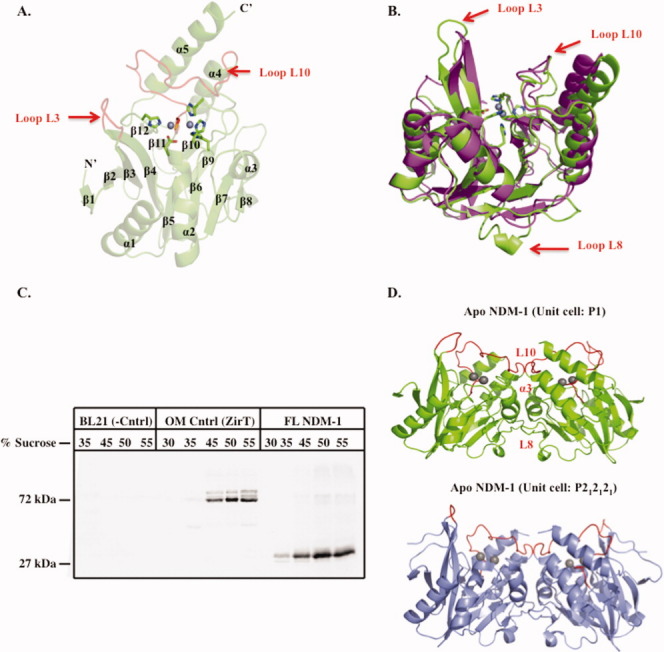
Overall structure of apo-NDM-1, NDM-1 localization, and crystallographic dimerization. A: Cartoon of apo-NDM-1 structure (green), active-site zinc ions (gray spheres), zinc ligands (stick, CPK coloring), and active-site loops L3 and L10 (red). Secondary structure designations are labeled. B: Superposition of NDM-1 on apo-VIM-2 (PDB ID 1K03)15 (magenta). Apo-NDM-1 zinc ligands are colored by atom, and the NDM-1 loops L3, L8, and L10 are highlighted with a red arrow. C: Western blot of a step sucrose gradient of membrane-associated 8xHIS FL-NDM-1. Negative control is E. coli BL21 membranes only. Positive control is outer membrane protein ZirT.16 D: NDM-1 crystallographic dimer interface for both apo (green) and ampicillinic acid-bound (gray, PDB ID: 3Q6X)13 forms. The L8, L10, and α3 helix constitute the dimer interface and are labeled on the apo structure (red).
