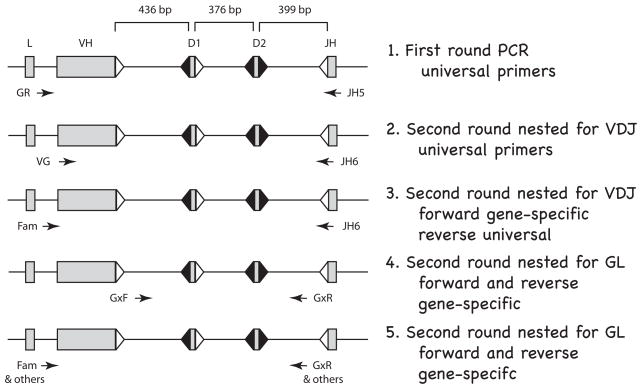
Figure 1.
Flow diagram for single-cell PCR in shark B cells. The rearranging gene segments (VH, D1, D2, JH) are separated by ~400 bp; the specific distances are from the gene G2A; L is leader. The flanking recombination signal sequences (RSS) with 23 bp spacers are represented as open triangles and RSS with 12 bp spacers filled triangles. Arrows indicate position and orientation of PCR primers. Line 1. First round PCR. A 40-cycle PCR was performed on B cells and on RBC, using “universal” primers detecting all functional IgH. Thereafter, the PCR products were depleted of oligonucleotides and used in the following four kinds of nested second-round reactions (25–30 cycles). Line 2. Second round nested for VDJ. Universal primers targeting FR1 and JH can amplify VDJ and/or GL fragments in B cells but only GL in the RBC samples. Line 3. Second round nested for VDJ. Specific forward primers reveal which subfamily (G1, G2, G3, G4, G5) the VDJ belongs to. VDJ is isolated and sequenced. Line 4. Second round nested for GL. Subfamily specific primers in the V-D1 and D2-J intersegmental regions amplify the non-rearranged genes. The five subfamily reactions are analyzed by restriction enzyme analyses detailed in Fig. S2, Supplementary Materials. Line 5. Second round nested for GL. The unrearranged VH is amplified for sequencing by using subfamily or gene-specific primers. Primer information is supplied in Materials and Methods.