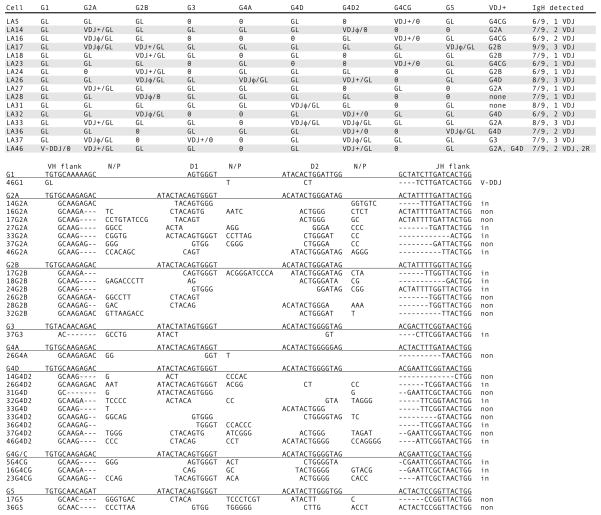
Figure 3.
Single-cell PCR results from shark-LA sIgM+ cells. Top. Rearrangement configurations of the nine IgH in each cell. The IgH genes present in shark-LA were: G1, G2A, G2B, G3, G4A, G4D, G4D2, G4CG, and G5. The status of the Ig gene segments is indicated as fully rearranged (VDJ), partly rearranged as V-DDJ, or not rearranged (GL). In-frame, potentially functional VDJ are indicated by plus (+) sign; nonproductive VDJ have φ sign. Zero indicates that the gene could not be detected. The column VDJ+ shows the probable IgH gene encoding the receptor. IgH detected indicates tally of the genes characterized per B cell and the number of unique VDJ. Bottom. CDR3 of VDJ. Their junctions are shown according to assignment to VH flank, D1 gene, D2 gene, and JH flank, classified according to subfamily. N/P, nontemplated or palindromic sequence. Dashes denote nucleotide deletions at the flanks. At the right, the VDJ are indicated as in-frame or nonproductive (non). V-DDJ is sequence with only two rearrangement (2R) events, where the V-D intersegmental region was not been deleted by recombination.