Abstract
The Notch signal transduction pathway mediates important cellular functions through direct cell-to-cell contact. Deregulation of Notch activity can lead to an altered cell proliferation and has been linked to many human cancers. Casein kinase 2 (CK2), a ubiquitous kinase, regulates several cellular processes by phosphorylating proteins involved in signal transduction, gene expression, and protein synthesis. In this report we identify NotchICD as a novel target of phosphorylation by CK2. Using mapping and mutational studies, we identified serine 1901, located in the ankyrin domain of Notch, as the target amino acid. Interestingly, phosphorylation of serine 1901 by CK2 appears to generate a second phosphorylation site at threonine 1898. Furthermore, threonine 1898 phosphorylation only occurs when Notch forms a complex with Mastermind and CSL. Phosphorylation of both threonine 1898 and serine 1901 resulted in decreased binding of the Notch-Mastermind-CSL ternary complex to DNA and consequently lower transcriptional activity. These data indicate that the phosphorylation of serine 1901 and threonine 1898 negatively regulates Notch function by dissociating the complex from DNA. This study identifies a new component involved in regulation of NotchICD transcriptional activity, reinforcing the notion that a precise and tight regulation is required for this essential signaling pathway.
Keywords: Notch Pathway, Protein-DNA Interaction, Protein Kinases, Protein Phosphorylation, Signal Transduction, Transcription Regulation, Hierarchical Phosphorylation, Transcriptional Activation Complex
Introduction
Notch signaling regulates several cellular processes such as cell proliferation, differentiation and apoptosis, thereby playing a key role in cellular homeostasis. There are four Notch family members that show complementary and combinatorial expression patterns depending on cell type (1, 2). Notch signaling is initiated by the interaction between the DSL ligand (Delta, Serrate, and Lag-2) and the extracellular domain of Notch, which is brought about by cell-to-cell contact. This interaction leads to proteolytic cleavage events that result in the release of Notch intracellular domain (NotchICD) from the plasma membrane, which then translocates into the nucleus (1, 3, 4). Through a stepwise assembly, NotchICD interacts with the DNA-binding protein CSL (for CBF1, Su(H), and Lag-1) and the co-activator Mastermind-like 1 (Maml1)4 (5–12). This interaction results in the formation of the transcriptional activation complex on DNA, which then regulates the expression of downstream target genes (10, 13, 15–20).
Inappropriate expression and deregulation of mammalian Notch is involved in the generation of neoplasia (21–29), indicating that precise spatiotemporal regulation of Notch signaling is required for proper cellular homeostasis. Several reports have shown that Notch is regulated by posttranslational modifications such as phosphorylation and ubiquitination during multiple steps in the signaling pathway (30–44). In particular, proteasome-mediated degradation of Notch after phosphorylation on the PEST domain is a key mechanism of regulating the Notch pathway (38, 39). In addition to phosphorylation within the PEST domain of NotchICD, recent evidence also suggests that phosphorylation occurs in other domains, and this has an effect on the Notch-driven transcriptional activity (45, 46) as well as its interactions with other proteins, such as Pin-1 (47). Although a general scheme for Notch signaling and its regulation has been identified, the role of post-translational modifications in general remains elusive (30). Identification and characterization of the post-translational modifications of Notch will aid in understanding the delicate balance of Notch activity regulation and the cross-talk of Notch with other signaling pathways.
In this study we report that NotchICD is phosphorylated by casein kinase 2 (CK2). CK2 phosphorylates NotchICD on serine 1901, which is located within the ankyrin repeat domain of NotchICD. Furthermore, in the transcriptional activation complex the site of phosphorylation is located in a loop predicted to be in close proximity with Maml1 and CSL. Interestingly, phosphorylation of Ser-1901 by CK2 generates a second phosphorylation site at threonine 1898 (Thr-1898). Our data suggest that the phosphorylation at Thr-1898 only occurs when Notch forms a complex with Maml1 and CSL. Phosphorylation of both Thr-1898 and Ser-1901 diminished the formation of a Notch-Maml1-CSL ternary complex on DNA and had a negative effect on the transcriptional activity of Notch. Our data indicate that CK2 negatively regulates Notch signaling by phosphorylating Ser-1901, which most likely leads to phosphorylation of Thr-1898 and subsequent destabilization of the NotchICD-Maml1-CSL activation complex on DNA.
EXPERIMENTAL PROCEDURES
DNA Constructs and Site-directed Mutagenesis
Wild type (wt) NotchICD and NotchICDΔ2202 were cloned in pCDNA3.1 (Invitrogen) as described in Jeffries and Capobianco (48). Point mutations were made using the QuikChange site-directed mutagenesis protocol (Stratagene) with primer pairs matching the target regions. Table 1 shows the sequences of the primers used for generating the mutants. For each mutation, two individual clones were picked and sequenced to confirm the mutation. Truncated forms of the mutants were generated by PCR using primers described previously (48) and cloned into pCDNA3.1 at BamHI and XhoI sites. For bacterial expression, mutants were subcloned into the pGEX4T-2 vector (Amersham Biosciences) to generate GST fusion proteins. The mutants were also subcloned into pBabe-puro and MIG (MCSV-IRES-GFP) retroviral vectors at BamHI and XhoI sites. Baculovirus expression constructs were generated as previously described (5).
TABLE 1.
Sequences of primers used for site-directed mutagenesis
The mutated sequences are underlined.
| Mutation | Primer sequences |
|---|---|
| Gly-1901 | 5′-ggcctggagacgggcaacggCgagGaAgaggagGACGCG-3′ |
| Ala-1901 | 5′-CTGGAGACGGGCAACGCCGAGGAAGAGGAGGAC-3′ |
| Asp-1901 | 5′-ggcctggagacgggcaacgacgaggaagaggaGgacgcg-3′ |
| Ala-1898 | 5′-GGCGGCCTGGAGGCCGGCAACAGCGAG-3′ |
| Ala-1912 | 5′-gcgccggccgtcatcgccgacttcatctaccag-3′ |
| Ala-1920 | 5′-ttcatctaccagggcgccgccctgcacaaccagaca-3′ |
| Ala-1925 | 5′-agcctgcacaaccaggcagaccgcacgggcgag-3′ |
| Ala-1928 | 5′-aaccagacagaccgcgcgggcgagaccgccttg-3′ |
Cell Culture and Transfection
293-T and H1299 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37 °C in 5.5% CO2. HC-11 cells were maintained in RPMI supplemented with 10% FBS, 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 5 μg/ml insulin, and 10 ng/ml EGF. For transfection, 4 × 106 cells were seeded in 10-cm tissue culture plates. Transfection was carried out with 8 μg of DNA and 20 μl of Lipofectamine reagent (Invitrogen). The cells were harvested for assay or labeled with [32P]orthophosphate 48 h post-transfection. Spodoptera frugiperda IPLB-Sf21 cells were maintained in Grace's Insect medium (Invitrogen) supplemented with 10% FBS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg of streptomycin per ml (Invitrogen).
Retroviral Infections
For generating retrovirus, 293-T cells were co-transfected with the retroviral vector and the SV40 psi- packaging vector. Virus was collected 24 and 48 h post-transfection and filtered using syringe filters. HC-11 was plated in 6-well plates (105 cells). The cells were infected the next day with the virus-containing medium in the presence of 8 μg/ml hexadimethrine bromide (Sigma). The cells were assessed for infection efficiency 72–96 h post-infection by GFP expression and harvested for RNA isolation.
In Vivo Orthophosphate Labeling and Immunoprecipitation
HeLa and RKE cells, transiently or stably expressing NotchICD and NotchICD variants, were cultured to 60–80% confluence in 10-cm culture plates and phosphate-starved for 2 h in phosphate-free medium (phosphate-free DMEM (Invitrogen) plus 10% dialyzed FBS (Invitrogen)) at 37 °C in 5.5% CO2. Cells were labeled with 3 mCi of [32P]orthophosphate (8500–9120 Ci/mmol) in 3 ml of phosphate-free medium for 4 h at 37 °C in 5.5% CO2. After labeling, the cells were washed with ice-cold PBS and harvested in Nonidet P-40 lysis buffer (150 mm NaCl, 50 mm HEPES, pH 7.4, 1.5 mm EDTA, 10% glycerol, 1% Nonidet P-40, supplemented with 1 mm DTT, 50 mm NaF, 0.5 mm sodium orthovanadate, and protease inhibitors (2 mm Pefabloc, 5 μg/ml leupeptin and 2 μg/ml aprotinin (Roche Applied Science)). To reduce nonspecific interactions, 0.2% SDS was added to the Nonidet P-40 lysis buffer. However, SDS was excluded in the co-immunoprecipitation studies.
NotchICD was immunoprecipitated using anti-c-Myc monoclonal antibody (9E10, Covance) or anti-Notch1 polyclonal antibody (927) followed by the addition of protein A-Sepharose beads (Sigma) for 1 h at 4 °C. After extensive washing with the Nonidet P-40 lysis buffer, the sample was resolved by SDS-PAGE and exposed to x-ray film. Alternatively, the gel was transferred to PVDF filter (Immobilon-P; Millipore) for Western blotting or phosphoamino acid analysis. For CNBr peptide mapping, proteins were transferred to nitrocellulose membrane (Schleicher & Schuell). Western blot analysis was performed using indicated antibodies and ECL protocol following the manufacturers' protocol (GE Healthcare).
Fractionation of Cell Lysate by Gel Filtration
3–6 × 107 RKE cells and 4–8 × 106 293-T cells were washed with PBS and resuspended in hypotonic lysis buffer (40 mm Tris-HCl, pH 7.4, 10 mm NaCl, 1 mm EDTA, 0.5 mm DTT, supplemented with protease inhibitor mixture). Cells were incubated on ice for 10 min and disrupted by Dounce homogenization. Nuclei were removed by low speed centrifugation (1600 × g) at 4 °C. The S-100 lysate was obtained by centrifuging the post nuclear lysate at 100,000 × g for 1 h at 4 °C. 200 μl of the S-100 lysate was fractionated by size exclusion chromatography in a FPLC using a Superose 6 10/300 GL (GE Healthcare) column equilibrated with column buffer (150 mm NaCl, 40 mm Tris-HCl, pH 7.4, 1 mm EDTA, 0.5 mm DTT, 0.001% Nonidet P-40).
Analysis of nuclear fractions for detecting NotchICD alone and in complex with Maml1 and CSL was carried out by transfecting 293-T with the appropriate combination of plasmids and further isolating the nuclei as described above. Nuclear fractions were run on a Superose 6 column, and the elution fractionate was then analyzed by SDS-PAGE and Western blot.
Expression of GST Fusion Proteins and in Vitro Labeling with [γ-32P]ATP
GST-NotchICD and GST-NotchICD variants were expressed in bacterial strain BL21 (Stratagene). Cells were grown at 37 °C in LB medium with 100 μg/ml ampicillin and induced for 3 h at 30 °C with 1.0 mm isopropyl 1-thio-β-d-galactopyranoside. Bacteria were collected by centrifugation at 5000 rpm for 10 min at 4 °C. Bacterial pellets were resuspended in 30 ml of ice-cold PBS (150 mm NaCl, 1.47 mm KH2PO4, 0.8 mm Na2HPO4) containing 2 m urea and disrupted by sonication in the presence of 1% Triton X-100. The lysate was centrifuged at 14,000 × g for 20 min at 4 °C. Glutathione-agarose beads (Sigma) were added to the lysate and incubated at room temperature for 1 h on a rotator. Beads were washed with PBS and re-suspended in an equal volume of PBS supplemented with 0.5 mm DTT and protease inhibitors and further stored at −70 °C.
In vitro labeling reactions were performed using purified GST fusion proteins bound to glutathione-agarose beads as a substrate and purified recombinant protein kinase CK2 catalytic subunit α (New England Biolabs) or FPLC-separated RKE cell lysate fractions as the kinase source. Each labeling reaction consisted of 10 μl of cell lysate fraction, 100 μm ATP, 5 μCi of [γ-32P]ATP (6000 mCi/mmol), 1 μg of GST protein bound to beads, 10 μl of 10× reaction buffer (100 mm Tris-HCl, pH 7.2, 10 mm MgCl2 supplemented with phosphatase inhibitors (50 mm NaF and 0.5 mm vanadate), 1 mm DTT, and protease inhibitors). Reactions were incubated at 37 °C for 1 h. Beads were washed with 1× reaction buffer and analyzed by SDS-PAGE. Samples were either stained with Coomassie Blue and visualized by autoradiography or transferred to PVDF membrane for Western blotting and phosphoamino acid analysis. For CNBr peptide mapping, proteins were transferred to nitrocellulose membrane.
To confirm that the kinase was CK2, a peptide substrate for CK2 (New England Biolabs) was also used as the substrate in the in vitro labeling reaction. The labeled product was then analyzed on a 24% low-bisacrylamide Tricine gel (49, 50).
Baculovirus Protein Purification
Recombinant CSL, NotchICD, and Maml1 were expressed and purified from Sf21 cells as previously described (5). Briefly, 3 × 107 Sf21 cells in 15-cm plates were infected at a multiplicity of infection of 2–5 with the baculovirus encoding the appropriate gene and harvested 48–72 h after infection. Cells were subsequently resuspended in high salt lysis buffer (500 mm NaCl, 40 mm Tris-HCl, pH 7.4, 1 mm EDTA, 10% glycerol, 0.02% Nonidet P-40, 1 mm DTT, supplemented with protease inhibitors) and lysed in a glass Dounce with 30 strokes of a pestle A over 30 min on ice. The lysates were centrifuged at 100,000 × g for 30 min at 4 °C. An equal volume of dilution buffer (40 mm Tris-HCl, pH 7.4, 10% glycerol, 0.02% Nonidet P-40, 1 mm DTT, and protease inhibitors) was then added to the supernatant. FLAG-tagged proteins were incubated with 100–200 μl of M2 FLAG-agarose beads (Sigma), and the His tagged proteins were incubated with 100–200 μl of nickel-nitrilotriacetic acid-agarose beads (Qiagen) for 3–5 h at 4 °C while rocking. The beads were washed in lysis buffer (150 mm NaCl, 40 mm Tris-HCl, pH 7.4, 1 mm EDTA, 10% glycerol, 0.02% Nonidet P-40, 1 mm DTT and protease inhibitors). FLAG-tagged proteins were eluted with elution buffer (150 mm NaCl, 40 mm Tris-HCl, pH 7.4, 0.4 mg/ml FLAG peptide (Sigma), 1 mm EDTA, 10% glycerol, 0.02% Nonidet P-40, 1 mm DTT and protease inhibitors). His-tagged proteins were eluted with elution buffer (250 mm imidazole, 40 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10% glycerol, 0.02% Nonidet P-40, 1 mm DTT, and protease inhibitors). All proteins were dialyzed in storage buffer (100 mm KCl, 20 mm HEPES, pH 7.9, 20% glycerol, and 1 mm DTT), separated into aliquots, and stored at −80 °C.
In-gel Kinase Assay and CNBr Analysis
For the in-gel kinase assays, GST or GST-NotchICDΔ2202 was polymerized in 12% SDS-polyacrylamide gels at the concentration of 50 μg/ml. Superose 6 fractionated lysates or purified CK2 catalytic subunit α was used as the kinase source. In-gel kinase reactions were performed as previously described (51).
CNBr analysis was performed by treating the protein on nitrocellulose membrane with 100 mg/ml CNBr in 70% formic acid. Samples were washed with distilled H2O, dried, and either loaded on 24% low bisacrylamide Tricine gels or 16.5% Tricine gels (Bio-Rad) and exposed to film (52).
Anti-phosphorylated Ser-1901 and Anti-phosphorylated Thr-1898 Antibody Preparation
Two different antibodies were generated to recognize NotchICD phosphorylated on Ser-1901 and Thr-1898. Peptides CLETGNpSEEEE and GGGLEpTGNSEEEEDC were used as antigens for antibody production in rabbit directed to recognize Ser-1901 and Thr-1898 phosphorylation, respectively. Both antibodies were affinity-purified first using non-phosphorylated peptides to remove antibodies not directed to the phosphorylated forms, and then the flow-through was affinity-purified using the phosphorylated peptides to end with the antibodies recognizing the phosphorylated forms. The entire procedure was performed by Open Biosystems.
Electrophoretic Mobility Shift Assay (EMSA) Experiments
EMSA was carried out by incubating 0.1 pmol of 5′-end-labeled double-stranded 1× CBF1 probe (5′-AAACACGCCGTGGGAAAAAATTTATG-3′), that has one high affinity binding site for CSL (underlined), with 50–200 ng of purified recombinant baculovirus proteins CSL, NotchICD, and Maml1 in EMSA buffer (100 mm KCl, 20 mm HEPES, pH 7.9, 0.2 mm EDTA, 1 mm DTT, 2 μg poly(dI·dC), 1 μg of BSA, 20% glycerol, and 0.25% Tween 20) in a final volume of 20 μl. After incubating for 40 min at room temperature, the DNA-protein complexes were separated on 5% non-denaturing polyacrylamide gels in 1× TGE buffer (0.25 m Tris, pH 8.3, 0.19 m glycine, 1 mm EDTA) at 4 °C) and visualized by autoradiography.
RT-PCR Analysis
The cells were harvested, and RNA was isolated using the RNAeasy kit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized using 1 μg of total RNA using Superscript Reverse Transcriptase (Invitrogen). 20 ng of RNA equivalent cDNA was used for PCR reactions. The quantitative PCR reactions were carried out on the Bio-Rad CFX96 using the SYBR Green PCR mix (Bio-Rad). The gene expression was normalized to hypoxanthine-guanine phosphoribosyltransferase (HPRT) expression.
Luciferase Reporter Assay
Luciferase assays were performed as described earlier (5). Basically, 6.0 × 104 H1299 cells were transfected with a total of 2 μg of DNA. Transfections included 0.4 μg of 8× CSL luciferase reporter vector (53), 0.4 μg of SV40 β-galactosidase (internal transfection control plasmid; Clontech), and 0.2 μg of the indicated NotchICD expression plasmid. The concentration of DNA was brought to 2 μg with pCDNA. Cells were transfected with FuGENE 6 (Roche Applied Science) and 48 h post-transfection, lysates were analyzed for luciferase activities using a Xylux Femtomaster FB 12 luminometer according to the manufacturer's suggested protocol (Promega). Luciferase values were corrected for transfection efficiency by normalizing to β-galactosidase activity.
Quantitative DNA Binding Assay
A 47-mer biotinylated dsDNA containing 2× CSL binding sites was bound to a 96-well streptavidin-coated plate (Greiner). The plate was washed 3 times before binding with 5× SSC (750 mm NaCl, 75 mm sodium citrate·2H2O, supplemented with 0.05% Tween 20) and post-binding with DNA binding buffer (100 mm Tris, pH 8, 150 mm NaCl, 1 mm DTT, 5 mm MgCl2, 0.1% Nonidet P-40, 5% glycerol, and 100 μg/ml bovine serum albumin). The second wash removes excess of unbound DNA. After washes, Notch (0.1 pmol) was preincubated on the plate in the presence or absence of 200 μm of ATP and increasing quantities of CK2 for 30 min before the addition of Maml-Luc (0.1 pmol) and CSL (1 pmol). Proteins were then incubated for 10 min at room temperature. Maml-Luc corresponded to a fusion between Maml1 (residues 1–305) and luciferase protein. After incubations, the plate was washed again three times with binding buffer to remove proteins that were not form part of a complex. Finally, luciferase assay reagent (LAR, Promega) was added, and luminescence was measured in a BMG plate reader.
RESULTS
NotchICD Is phosphorylated within the Ankyrin Domain by Casein Kinase 2
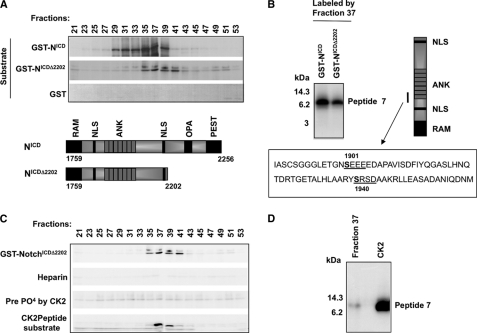
To identify novel kinases that phosphorylate NotchICD and regulate its function, we performed kinase assays with fractionated lysates from RKE cells transformed with NotchICD. Superose-6 FPLC-separated fractions were used as a kinase source, and purified GST-NotchICD and GST-NotchICDΔ2202 were used as substrates for in vitro kinase assays. GST-NotchICDΔ2202, a C-terminal deletion mutant that harbors the minimal region needed for cellular transformation (48), was used to identify kinases that regulate NotchICD function but do not phosphorylate the PEST domain. A kinase activity that peaked in fraction 37 phosphorylated both GST-NotchICD and GST-NotchICDΔ2202 (Fig. 1A). CNBr cleavage analysis of the labeled GST-NotchICD and GST-NotchICDΔ2202 was carried out to map the phosphorylation sites, and as shown in Fig. 1B, fraction 37 specifically labeled the ankyrin domain, and the target amino acid was on peptide 7 (Fig. 1B).
FIG. 1.
Identification of a kinase activity in NotchICD-transformed RKE cells. A, kinase assays were performed using FPLC-separated fractions of NotchICD-transformed RKE cell S100 lysate as the kinase source and purified GST-NotchICD or NotchICDΔ2202 as substrates. GST was used as a negative control. ANK, ankyrin; NLS, nuclear localization signal. B, CNBr cleavage analysis of fraction 37-labeled GST-NotchICD and GST- NotchICDΔ2202 is shown. Below, the amino acid sequence of peptide 7 shows putative phosphorylation sites. Potential CK2 sites are underlined, and the consensus sequence is shown. C, biochemical properties of fraction 37 kinase resemble that of CK2. In vitro kinase reaction shows labeling of GST-NotchICDΔ2202 by fraction 37 kinase. Heparin, a specific inhibitor of CK2, also inhibits fraction 37 kinase activity in in vitro kinase reactions. Prephosphorylation of GST-NotchICDΔ2202 by CK2 inhibits the phosphorylation of GST-NotchICDΔ2202 by the fraction 37 kinase. The fraction 37 kinase also phosphorylates the CK2 peptide substrate (RRREEETEEE) in in vitro kinase reactions. D, CNBr cleavage analysis of the fraction 37 kinase and CK2-labeled GST-NotchICDΔ2202 shows that peptide 7, residing in the ankyrin domain, is targeted by both kinases.
Analysis of the amino acid sequence of peptide 7 revealed two serine residues, Ser-1901 and Ser-1940, within peptide 7 of NotchICD that fit the consensus sequence for casein kinase 2 ((T/S)XX(E/D)) (Fig. 1B). This prompted us to investigate if the kinase in fraction 37 is CK2. To confirm the identity of this kinase, it was first tested whether heparin, a known inhibitor of CK2 activity, could block fraction 37 kinase activity. An in vitro kinase reaction demonstrated that the kinase activity that peaked in fraction 37 was inhibited in the presence of heparin (Fig. 1C). Next we tested if fraction 37 kinase could also phosphorylate a CK2 peptide substrate (RRREEETEEE), showing that both fraction 37 kinase and CK2 had similar substrate specificity (Fig. 1C). Finally, to demonstrate that the site(s) of phosphorylation is the same for fraction 37 kinase and CK2, we prephosphorylated GST-NotchICDΔ2202 with recombinant CK2 using unlabeled ATP as phosphate donor. The prephosphorylated GST-NotchICDΔ2202 was then used as a substrate for fraction 37 kinase in an in vitro kinase reaction. Prephosphorylation of GST-NotchICDΔ2202 by CK2 prevented incorporation of 32P by fraction 37 kinase (Fig. 1C). This suggests that fraction 37 kinase and CK2 likely target the same Ser residue(s) on peptide 7. CNBr cleavage analysis of GST-NotchICDΔ2202 phosphorylated by fraction 37 and recombinant CK2 confirmed that the target Ser was on peptide 7 (Fig. 1D).
Ser-1901 within the Ankyrin Domain of NotchICD Is the Target for CK2
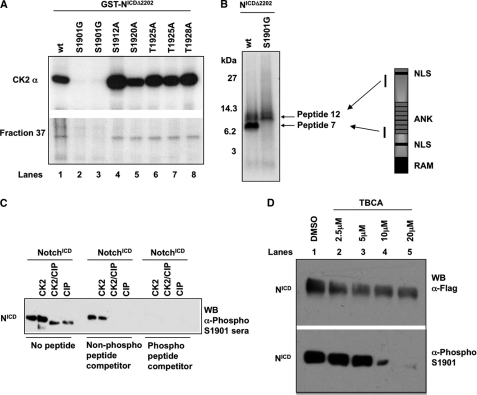
Peptide 7 contains CK2 consensus sequence (T/S)XX(E/D) at Ser-1901. To determine if Ser-1901 was a target of CK2, we changed Ser-1901 to Gly by site-directed mutagenesis and constructed GST-NotchICDΔ2202 S1901G. As controls, several other Ser and Thr residues in peptide 7 were also mutated. In vitro kinase assays using GST-NotchICDΔ2202 S1901G as substrate demonstrated that mutation of Ser-1901 to Gly prevented phosphorylation by both the CK2 and fraction 37 kinase (Fig. 2A, lane 2 and 3). In contrast, mutation of other Ser and Thr residues to Ala did not affect phosphorylation by either CK2 or fraction 37 kinase (Fig. 2A, lanes 4–8). These results demonstrate that Ser-1901 in NotchICD is the target of CK2 and fraction 37 kinase in vitro.
FIG. 2.
Ser-1901 in the ankyrin domain of NotchICD is the CK2 phosphorylation site. A, in vitro kinase assays were performed with CK2 and fraction 37 as kinase sources and GST-NotchICDΔ2202 and several point mutants of Notch as substrate. B, CNBr cleavage analysis of in vivo labeled NotchICDΔ2202 and NotchICDΔ2202 S1901G shows that peptide 7 phosphorylation is absent in NotchICDΔ2202 S1901G. NLS, nuclear localization signal; ANK, ankyrin. C, Ser-1901 phospho-specific antibody was tested on purified NotchICD either phosphorylated by CK2 or dephosphorylated by alkaline phosphatase (CIP). Non-phospho and phospho peptides were used to block the antibody. D, 293-T cells were transfected with NotchICD and treated with either DMSO or with increasing concentrations of the CK2 inhibitor TBCA for 24 h. 100 μg of total cell extract was analyzed for NotchICD expression with anti-FLAG antibody, and phosphorylation of Ser-1901 was detected using the phospho-specific Ser-1901 antibody. WB, Western blot.
To determine if 1901 is a phosphorylation site for CK2 in vivo as well, we transiently expressed NotchICDΔ2202 and NotchICDΔ2202 S1901G in HeLa cells and performed in vivo labeling. NotchICDΔ2202 and NotchICDΔ2202 S1901G were immunoprecipitated from cell lysates using anti-NotchICD and subjected to CNBr cleavage. Both NotchICDΔ2202 and NotchICDΔ2202 S1901G were phosphorylated on peptide 12, but peptide 7 was only detected in NotchICDΔ2202 and not in NotchICDΔ2202 S1901G (Fig. 2B). Furthermore, the fact that mutation of Ser-1901 to Gly completely prevented the phosphorylation of peptide 7 in NotchICDΔ2202 S1901G indicates that Ser-1901 was the only accessible phosphorylation site in peptide 7. In contrast, Ser-1940 is not phosphorylated by CK2 despite the fact that it fits the CK2 consensus sequence. Taken together, these data indicate that Ser-1901 is phosphorylated in cells with a high degree of specificity and that the kinase responsible is most likely CK2.
To further demonstrate that Ser-1901 of NotchICD is phosphorylated in vivo, we generated an antibody against phosphorylated Ser-1901. Fig. 2C demonstrates specificity of the antibody in recognizing recombinant NotchICD phosphorylated at Ser-1901 by CK2. In particular, antibody binding was out-competed by a specific phosphorylated peptide competitor but not by a non-phosphorylated peptide competitor (Fig. 2C).
To demonstrate that our antibody specifically detects phosphorylation of Ser-1901 by CK2, we treated 293-T cells transfected with NotchICD with the specific CK2 inhibitor, TBCA (54). Increasing the concentration of TBCA resulted in NotchICD no longer being phosphorylated at Ser-1901 (Fig. 2D, lanes 2–5). However, cells treated with DMSO expressed NotchICD that was phosphorylated at Ser-1901 (Fig. 2D, lane 1). As a control for NotchICD expression, cells treated with either TBCA or DMSO expressed similar levels of NotchICD (Fig. 2D, lanes 1–5, upper panel).
NotchICD Phosphorylation on Ser-1901 Occurs Independent of the Association with Maml1 and CSL
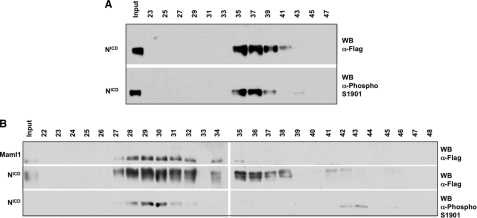
The ankyrin repeat domain of NotchICD is located in the central region of the protein C-terminal to the RAM domain. This domain, as seen in the crystal structure of the activation complex, likely serves as a scaffold function for NotchICD activity by interacting simultaneously with Maml1 and CSL (55, 56). Several reports have pointed out the indispensable role of this domain in transactivation as determined by deletion and mutation analysis (6, 10, 48, 57–61). Phosphorylation of NotchICD by CK2 within the ankyrin domain raises the possibility that phosphorylation may affect NotchICD activity by modulating the functional interactions between NotchICD and other proteins, such as Maml1 and CSL. This idea is reinforced by observing the crystal structure of the Notch transcriptional activation complex (see Fig. 8A), which reveals that the loop containing Ser-1901 is in close proximity to Maml1 and CSL. To determine if NotchICD phosphorylated at Ser-1901 is associated with Maml1 and CSL, 293-T cells were transfected with either NotchICD alone or with NotchICD, Maml1, and CSL. 48 h post-transfection, nuclear lysate was fractionated by size exclusion chromatography on a Superose 6 column. NotchICD, which is not associated with Maml1 and CSL, appears in fraction 37 and is phosphorylated on Ser-1901 as detected by the phospho-specific antibody (Fig. 3A). In the presence of Maml1 and CSL, phosphorylated NotchICD was only detected in fractions 30 and 31, which corresponds to the complex with Maml1 and CSL (Fig. 3B) (53). Taken together, these results suggest that even though Notch phosphorylated on 1901 is preferentially associated with Maml1 and CSL, phosphorylation on Ser-1901 take place independent and before complex formation, which is consistent with CNBr-mapping results of HeLa cells transfected with NotchICD (Fig. 2B). Therefore, the phosphorylation does not dissociate the interaction with these proteins in the ternary (higher molecular weight) complex. These experiments, however, do not address the role of this phosphorylation on complex formation on DNA and subsequent transcriptional activation.
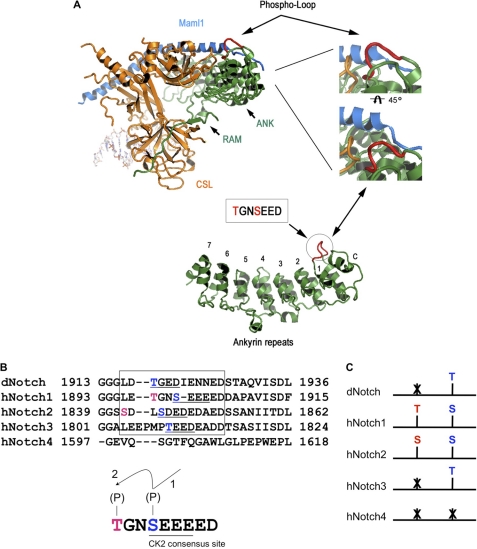
FIG. 8.
Location of the phosphoregulatory loop in the NotchICD-Maml1-CSL complex on DNA. A, a ribbon representation of the activation complex on DNA shows the phospho-loop. Representation was prepared in PyMOL (14) using the coordinates from the crystal structure of the quaternary ANK/RAM, Mastermind, CSL, and DNA complex (PDB 2FO1). RAM and ankyrin repeat domains of Notch are shown in green, CSL in orange, and Mastermind is in light blue. The phosphoregulatory loop in Notch is denoted by an arrow and shown in red. Top left, the structure of the activation complex on DNA is shown. Top right, zoom shows the phospho-loop in proximity with Mastermind and CSL. Bottom, the position of the phospho-loop relative to the ankyrin domain of Notch is shown. CK2 consensus sequence is highlighted in a box next to the red circle denoting the phospho-loop. B, amino acid alignment of four human Notch (hNotch1–4) proteins and the Drosophila Notch Protein (dNotch) reveals a conserved CK2 consensus sequence for human Notch1–3 and Drosophila Notch (underlined). Blue and red highlights denote residues that might be equivalent to human Notch1 Ser-1901 and Thr-1898, respectively. Below is a schematic showing hierarchical phosphorylation event in which Ser-1901 is first phosphorylated by CK2 followed by phosphorylation of Thr-1898. C, schematics show the phosphorylated residues in hNotch1 and their predicted equivalents in other Notch paralogs.
FIG. 3.
Ser-1901 is phosphorylated when NotchICD is alone or associated with Maml1 and CSL. A, 293-T cells were transfected with NotchICD, and the nuclear fraction was run on a Superose 6 column. Fractions were analyzed either by phospho-specific Ser-1901 antibody or by anti-FLAG antibody. B, 293-T cells were transfected with NotchICD, Maml1, and CSL, and the nuclear fraction was run on a Superose 6 column. Elution fractions were analyzed either by phospho-specific Ser-1901 antibody or anti-FLAG antibody. WB, Western blot.
CK2 Phosphorylation of NotchICD at Ser-1901 Decreases the Notch Activation Complex Binding to DNA in Vitro
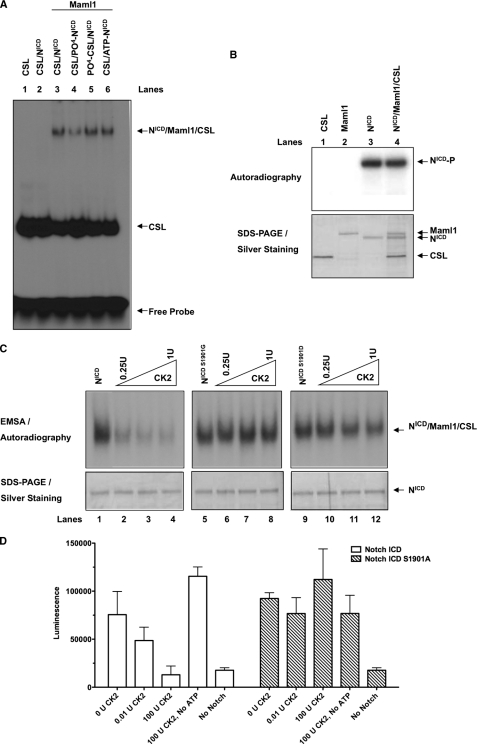
To determine the effect of the phosphorylation at Ser-1901 on the ability of the active complex to bind DNA, EMSA analyses were performed with recombinant baculovirus-expressed proteins NotchICD, Maml1, and CSL. Results demonstrated that in the presence of NotchICD, Maml1, and CSL, an activation complex formed on DNA (Fig. 4A, lane 3). Interestingly, prephosphorylation of NotchICD by CK2 resulted in a decrease in the DNA-bound activation complex (Fig. 4A, lane 4). However, the CSL-DNA complex was not affected (Fig. 4A, compare lanes 2, 3, and 4), indicating that phosphorylation affected only the NotchICD-Maml1-CSL complex. Furthermore, preincubation of CSL with CK2 or preincubation of NotchICD with ATP in the absence of CK2 had no effect on the formation of the activation complex on DNA (Fig. 4A, lanes 5 and 6), which demonstrates specificity.
FIG. 4.
Phosphorylation of Ser-1901 by CK2 affects the NotchICD-Maml1-CSL complex on DNA. A, EMSAs were performed on the indicated combinations of proteins (100 ng each). Proteins were incubated with 32P-labeled 1× CBF1 oligonucleotide containing a CSL consensus binding site, resolved on a 5% nondenaturing gel, and visualized by autoradiography. PO43−-NotchICD indicates NotchICD prephosphorylated by CK2 (0.25 units/100 ng of protein), PO43−-CSL indicates CSL treated with CK2 (0.25 units/100 ng protein), and ATP-NotchICD indicates NotchICD preincubated with ATP in the absence of CK2. B, in vitro CK2 assays were performed using baculovirus-expressed recombinant proteins as substrate and [γ-32P]ATP as phosphate donor. Samples were separated by SDS-PAGE. Autoradiography was done to detect 32P-labeled proteins (upper panel), and the same gel was silver-stained to show relative amounts of protein loaded in each lane (lower panel). C, increasing the amount of CK2 (0.25–1.00 units/100 ng of protein) did not affect the NotchICDS1901G-Maml1-CSL complex from binding to DNA as seen by EMSA. The experiment was performed in similar manner as A but with increasing concentrations of CK2. D, quantitative DNA binding assay was performed as described under “Experimental Procedures” with 2× CSL oligo in the presence of NotchICD wt or S1901A and increasing concentrations of CK2. Incubating with CK2 reduced the luciferase activity of the wt NotchICD but had no effect on S1901A. Incubating NotchICD wt with CK2 in the absence of ATP had no inhibitory effect on complex binding to DNA. U, unit(s).
To determine if the decrease in activation complex binding to DNA was a result of either Maml1 or CSL being phosphorylated by CK2, in vitro kinase assays were performed using purified NotchICD, Maml1, and CSL. Alone or in complex with Maml1 and CSL, NotchICD was phosphorylated by CK2 (Fig. 4B, lanes 3 and 4). In contrast, Maml1 and CSL were not phosphorylated by CK2 either alone or in complex with NotchICD (Fig. 4B, lanes 1, 2, and 4). These data indicate that CK2 specifically phosphorylates NotchICD and not Maml1 and CSL.
To determine if the decrement in the activation complex binding to DNA was specifically due to phosphorylation at Ser-1901 by CK2, NotchICD proteins containing S1901G and S1901D mutations were expressed in SF21 cells and affinity-purified. Recombinant proteins, NotchICD, NotchICD S1901G, and NotchICD S1901D, were prephosphorylated with increasing amounts of CK2, and activation complex binding to DNA was analyzed by EMSA. Results demonstrated that increasing the amounts of CK2 decreased the amount of the wt NotchICD activation complex binding to DNA in a dose-dependent manner (Fig. 4C, left panel). In contrast, increasing the amount of CK2 had no apparent effect on the binding of the NotchICD S1901G activation complex to DNA (Fig. 4C, middle panel), indicating that phosphorylation of Ser-1901 has a negative effect on NotchICD complex binding to DNA. However, in the case of NotchICD S1901D, the presence of low levels of CK2 showed no noticeable decrease when compared with the mutant and the wt in the absence of CK2 (Fig. 4C, lanes 1, 9, and 10). Thus, NotchICD S1901D does not seem to function as a phosphomimetic.
To have a more quantitative estimation of the effect of Ser-1901 phosphorylation on the ability of the Notch complex to bind DNA, complex stability on DNA was further tested by a quantitative DNA binding assay. Briefly, a biotinylated oligo containing two CSL binding sites was immobilized on a 96-well streptavidin-coated plate. This was then incubated with a Maml1-luciferase fusion protein in combination with other proteins as indicated in Fig. 4D. Interaction of NotchICD-Maml1-CSL complex on DNA was measured by luciferase activity. With increasing concentrations of CK2, the ability of NotchICD complex to bind DNA showed a marked reduction. However, the phosphorylation-deficient (S1901A/G) mutant showed no change in the luciferase activity in the presence of CK2. Taken together, these results demonstrate that phosphorylation of NotchICD by CK2 negatively regulates the stability of the activation complex on DNA.
Thr-1898 Becomes a Second Phosphorylation Site in NotchICD
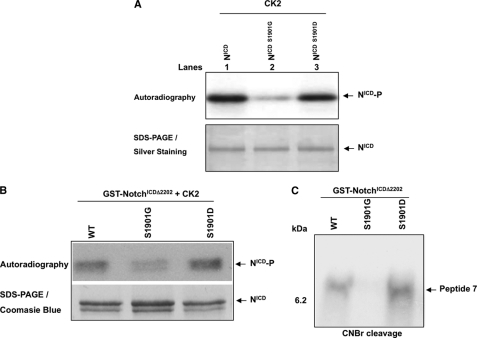
The observation that in the presence of high levels of CK2 there is a slight decrease on the binding of the NotchICD S1901D complex to DNA (Fig. 4C, lanes 11 and 12) led us to consider the possibility that the negative charge of the Ser to Asp substitution causes another residue to become the target of phosphorylation and presents a similar effect. To address if a second residue was being phosphorylated, in vitro kinase assays were performed with NotchICD, NotchICD S1901G, and NotchICD S1901D (Fig. 5A). As was previously observed, NotchICD was phosphorylated by CK2 (Fig. 5A, lane1), and in contrast, NotchICD S1901G was not (Fig. 5A, lane 2). Surprisingly, NotchICD S1901D was also phosphorylated by CK2 (Fig. 5A, lane 3), indicating that a new residue other than Ser-1901 was phosphorylated. Because Asp is unlikely to be phosphorylated, we analyzed the primary amino acid sequence of NotchICD and found that the substitution of Ser-1901 to Asp generates a new CK2 consensus site (TXXD) and, hence, identifies Thr-1898 as the candidate for the second phosphorylation. To determine if this new phosphorylated residue was also located in peptide 7, in vitro kinase assays were performed with GST-NotchICDΔ2202, GST-NotchICDΔ2202 S1901G, and GST-NotchICDΔ2202 S1901D (Fig. 5B) followed by CNBr cleavage analysis (Fig. 5C). Results revealed that both GST-NotchICDΔ2202 and GST-NotchICDΔ2202 S1901D were phosphorylated on peptide 7. As expected, GST-NotchICDΔ2202 S1901G was not phosphorylated on peptide 7 (Fig. 5C). These data indicate that a second residue, likely Thr-1898, becomes accessible for phosphorylation only when there is negative charge on 1901, namely when it is phosphorylated or when it is mutated to Asp, and further suggests the presence of a hierarchical phosphorylation event, which is demonstrated in subsequent experiments (see Fig. 7).
FIG. 5.
Phosphorylation of Ser-1901 by CK2 results in Thr-1898 being phosphorylated. A, shown is in vitro CK2 kinase assay using recombinant proteins NotchICD, NotchICD S1901G, and NotchICD S1901D as substrates. Autoradiography shows NotchICD and NotchICD S1901D are phosphorylated by CK2, whereas NotchICD S1901G is not labeled (upper panel). The same gel was silver-stained to show equal loading of the proteins (lower panel). B, in vitro CK2 kinase assays were performed on GST-NotchICDΔ2202, GST-NotchICDΔ2202 S1901G, and GST-NotchICDΔ2202 S1901D. Samples were separated by SDS-PAGE. Autoradiography was done to detect 32P-labeled proteins (upper panel). The same gel was Coomassie-stained to show relative amounts of protein loaded in each lane (lower panel). C, CNBr cleavage analysis of CK2-labeled GST-NotchICDΔ2202, GST-NotchICDΔ2202 S1901G, and GST-NotchICDΔ2202 S1901D shows GST-NotchICDΔ2202 and GST-NotchICDΔ2202 S1901D are phosphorylated within peptide 7.
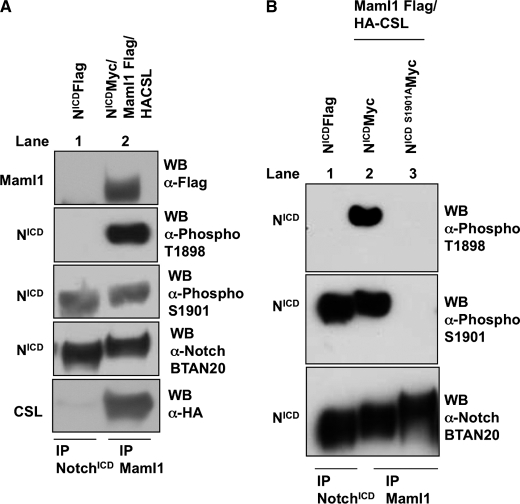
FIG. 7.
Phosphorylation of Thr-1898 is only detected when Notch forms a complex with Maml1 and CSL and requires phospho-priming of Ser-1901. A, 293-T cells were transfected either with NotchICD alone or NotchICD co-transfected with Maml1 and CSL cDNA constructs. 48 h post-transfection cells were harvested, and whole cell lysates were made. Anti-FLAG antibody was used either to immunoprecipitate (IP) NotchICD alone or NotchICD associated with Maml1 and CSL. Western blots (WB) were performed using anti-phospho Thr-1898 antibody to detect phosphorylated Thr-1898, anti-phospho Ser-1901 antibody to detect phosphorylated Ser-1901, anti-Notch antibody (BTAN20) to detect NotchICD, anti-Myc (9E10) antibody to detect Maml1, and anti-HA antibody to detect CSL. B, 293-T cells were transfected either with NotchICD alone or NotchICD and NotchICD S1901A in combination with Maml1 and CSL cDNA constructs. 48 h post-transfection cells were harvested, and whole cell lysates were made. Anti-FLAG antibody was used to immunoprecipitate NotchICD alone and NotchICD and NotchICDS1901A associated with Maml1 and CSL. Western blots were performed using anti-phospho-Thr-1898 antibody to detect phosphorylated Thr-1898, anti-phospho Ser-1901 antibody to detect phosphorylated Ser-1901, and anti-Notch antibody (BTAN20) to detect NotchICD. HA-CSL, hemagglutinin tagged CSL.
Phosphorylation on Ser-1901 and Thr-1898 of NotchICD Negatively Regulates DNA Binding and Transcriptional Activity
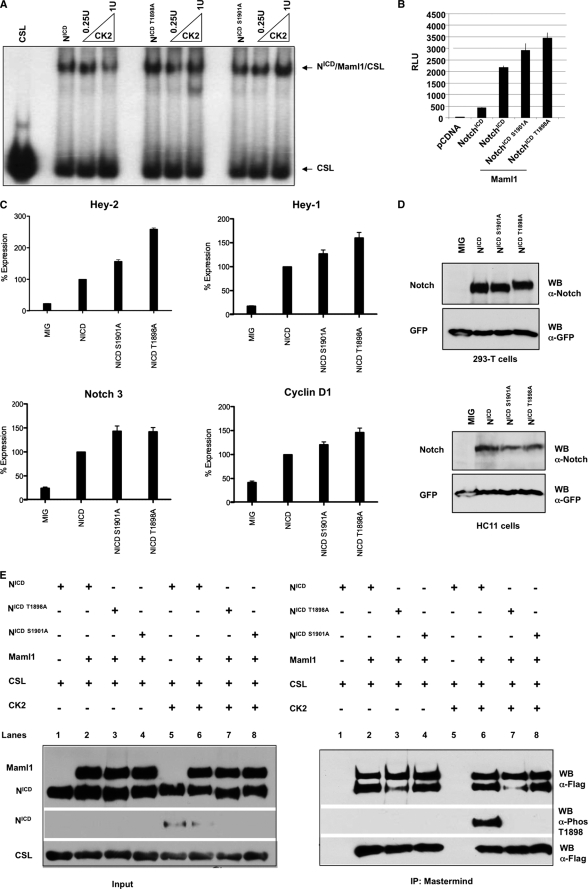
Because phosphorylating Ser-1901 likely leads to Thr-1898 being phosphorylated, this raises the possibility that phosphorylating Ser-1901 alone or both Ser-1901 and Thr-1898 reduces the stability of the activation complex on DNA. To test this, EMSA analysis was performed with purified NotchICD, NotchICD S1901A, and NotchICD T1898A phosphorylated by CK2 (Fig. 6A). As previously observed, prephosphorylating NotchICD affected the binding of the activation complex on DNA (Fig. 6A, left panel). Interestingly, preventing the phosphorylation of either Thr-1898 or Ser-1901 resulted in the activation complex binding to DNA (Fig. 6A, right panel), suggesting that both phosphorylation events are essential for decreasing binding of the Notch complex to DNA; the first phosphorylation event at Ser-1901, as it is needed to open the second site for phosphorylation, and the second phosphorylation event at Thr-1898, as it is likely to be the one that has the ultimate effect on the complex dissociating from DNA. The presence of DNA binding of NotchICD T1898A complex when CK2 is present, even though Ser-1901 is phosphorylated under these conditions, supports the idea that Thr-1898 phosphorylation is responsible for dissociating the complex from DNA (Fig. 6A).
FIG. 6.
Phosphorylation of Ser-1901 and Thr-1898 by CK2 is required to inhibit NotchICD activity. A, electrophoretic mobility shift assays were performed on NotchICD, NotchICD S1901A, and NotchICD T1898A (100 ng each) with equal molar quantities of Maml1 and CSL. Proteins were incubated with 32P-labeled 1× CBF1 oligonucleotide, resolved on a 5% nondenaturing gel, and visualized by autoradiography. Increasing the amount of CK2 (0.25–1.00 unit/100 ng of protein) did not affect the NotchICD S1901A-Maml1-CSL and NotchICD T1898A-Maml1-CSL complexes from binding DNA. B, H1299 cells were transfected with 0.05 μg each of NotchICD wt and mutant constructs and 0.1 μg of Maml1. 48 h post-transfection, cells were lysed, luciferase assays were performed in triplicate, and the results are shown in relative luciferase units (RLU). Error bars represent S.D. C, HC-11 cells were infected with retrovirus expressing each of the mutants indicated. 72–96 h post-infection, cells were harvested for RNA, and RT-PCR analysis was carried out for Hey-1, Hey-2, Cyclin-D1, and Notch 3 expression. The expression was normalized to hypoxanthine-guanine phosphoribosyltransferase expression. The experiment shown is representative of three independent experiments. Columns represent the mean, and bars denote S.D. within replicates of a single experiment. MIG, MCSV-IRES-GFP. NICD, Notch intracellular domain. D, top panel, retroviral vectors directed expression of Notch ICD wt and mutant proteins to similar levels in 293-T cells. Shown are Notch and linked GFP expression for normalization. Bottom panel, retroviral-infected HC11 cells show comparable expression levels of Notch wt and mutant proteins. WB, Western blot. E, immunoprecipitation (IP) assays were performed using different combinations of Baculovirus expressed Maml1, CSL and NotchICD, NotchICD T1898A, and NotchICDS1901A either phosphorylated or not phosphorylated by CK2. Immunoprecipitations were performed using anti-Maml1 antibody, and NotchICD, NotchICD T1898A, NotchICDS 1901A, and CSL were detected using anti-FLAG antibody. Phosphorylation of Thr-1898 was detected using anti-phospho-Thr-1898 antibody.
As described above, phosphorylation of Notch on Ser-1901 and Thr-1898 decreases the binding of the Notch activation complex to DNA. We decided to address if the decrease in DNA binding translates to a lower NotchICD transcriptional activity. For that purpose luciferase reporter assays were performed using an 8× CSL reporter in the presence of NotchICD wt or phosphorylation mutants. Results showed that both S1901A and T1898A resulted in higher luciferase activity compared with wt (Fig. 6B). These data suggest that CK2 negatively regulates Notch transcriptional activity by phosphorylating Ser-1901 and subsequently Thr-1898.
The results obtained suggested that phosphorylation of these sites may also have an effect on Notch target gene expression. To address endogenous gene expression, NotchICD, NotchICD S1901A, and NotchICD T1898A were introduced into HC-11 cells, and the expression of the Notch target genes Hey-1, Hey-2, Cyclin-D1, and Notch-3 were assessed by quantitative RT-PCR. Both S1901A as well as T1898A mutants displayed increased expression of the target genes compared with wild type (Fig. 6C). Specifically, in the case of Hey-2, NotchICD S1901A and NotchICD T1898A showed an increase of target gene expression of 57 and 160% when compared with the wt, respectively. However, expression levels of Notch wt and phosphorylation mutants were comparable (Fig. 6D). In contrast, deletion of the PEST domain of Notch results in dramatic increase in protein levels with a concomitant increase in gene transcription (∼280% increase in Hey-2 expression; data not shown). Collectively, these results demonstrate that phosphorylation of these residues reduce the active complex on DNA and have a negative effect on Notch mediated gene transcription and not protein stability.
Phosphorylation of Ser-1901 and Thr-1898 Does Not Dissociate the NotchICD-Maml1-CSL Protein Complex
Decreasing active complex stability on DNA can result from destabilizing the interactions between NotchICD, Maml1, and CSL. To determine if activation complex formation is inhibited by CK2 phosphorylating Ser-1901 and Thr-1898, co-immunoprecipitation assays were performed with purified Maml1 and CSL incubated with NotchICD, NotchICD T1898A, and NotchICD S1901A that were either phosphorylated or not phosphorylated by CK2 (Fig. 6E). When Maml1 was immunoprecipitated, prephosphorylation of NotchICD, NotchICD T1898A, and NotchICD S1901A by CK2 did not inhibit the formation of activation complexes (Fig. 6E, right panel, lanes 6–8). To determine if NotchICD phosphorylated at Thr-1898 was associated with Maml1 and CSL, Western blots were performed using an antibody raised against phosphorylated Thr-1898 (Fig. 6E, right panel). When Maml1 was immunoprecipitated, NotchICD phosphorylated at Thr-1898 and CSL co-immunoprecipitated (Fig. 6E, right panel, lane 6). As a negative control, CSL and NotchICD phosphorylated by CK2 in the absence of Maml1 were not immunoprecipitated by Maml1 antibody (Fig. 6E, right panel, lane 5). These results suggest that the phosphorylation at either or both of these residues does not have an observable effect on the stability of the NotchICD-Mastermind-CSL protein complex interactions.
Phosphorylation of Thr-1898 Follows Phosphorylation of Ser-1901 and Occurs When NotchICD Forms a Complex with Maml1 and CSL
To determine when NotchICD is phosphorylated at Thr-1898 in cells, 293-T were transfected either with NotchICD alone or NotchICD co-transfected with Maml1 and CSL (Fig. 7). When NotchICD was immunoprecipitated in the absence of exogenous Maml1 and CSL, phosphorylated Thr-1898 was not detected (Fig. 7A, lane 1, second panel). However, when Maml1 was immunoprecipitated to isolate NotchICD associated with Maml1 and CSL, phosphorylation at Thr-1898 was detected (Fig. 7A, lane 2, second panel). In both instances phosphorylation at Ser-1901 was observed (Fig. 7A, lanes 1 and 2, third panel). Western blot analysis with anti-Notch antibody was used to demonstrate equal immunoprecipitations of NotchICD between samples (Fig. 7A, lanes 1 and 2, fourth panel). Collectively, our data indicate that unlike the phosphorylation event at Ser-1901, phosphorylation at Thr-1898 only occurs when NotchICD associates with Maml1 and CSL.
To determine if Ser-1901 must be phosphorylated before phosphorylation of Thr-1898, 293-T were transfected either with NotchICD or NotchICD S1901A and co-transfected with Maml1 and CSL (Fig. 7B). When Maml1 was immunoprecipitated to isolate NotchICD associated with Maml1 and CSL, phosphorylation at Thr-1898 was not detected with the NotchICD S1901A mutant (Fig. 7B, lane 3). These data indicate that Thr-1898 phosphorylation occurs in cells and is dependent on phospho-priming at Ser-1901.
DISCUSSION
To date several laboratories including ours have demonstrated that NotchICD is subjected to multiple phosphorylations that modulate its transcriptional activity by regulating its stability or localization (31–46). Studies demonstrated that phosphorylation of the PEST domain by CDK8 targets NotchICD for ubiquitination and subsequent degradation (38, 39). GSK-3β has also been shown to phosphorylate NotchICD and protect it from proteasome-mediated degradation (35). More recently, it has been demonstrated that Notch is phosphorylated by Akt, which interferes with nuclear localization of NotchICD and, therefore, results in reduced transcriptional activity (44). Also, evidence over the last couple of years has shown that phosphorylation of NotchICD can directly affect its transcriptional activity (45, 46). One way that active complex formation on DNA could be affected is by phosphorylating and subsequently modifying the protein interactions of the ankyrin repeats of NotchICD with other components, such as CSL and Maml1. In particular, Down syndrome-associated kinase DYRK1A has been shown to phosphorylate NotchICD on the ankyrin repeats and results in attenuation of transcriptional activity (45). Similarly, Nemo-like kinase also phosphorylates NotchICD in the region C-terminal to the ankyrin repeat domain and reduces its transcriptional activity (46). In an effort to understand the role of phosphorylation in NotchICD function and identify kinase(s) responsible for regulating this activity, we carried out experiments that identified Notch as a substrate for CK2 and identified novel phosphorylation sites within the ankyrin repeats of NotchICD. We also demonstrate a hierarchical phosphorylation of NotchICD, where Ser-1901 is phosphorylated by CK2 followed by a second phosphorylation at Thr-1898. We further demonstrate that the phosphorylation of Thr-1898 only occurs when Notch forms a complex with Maml1 and CSL. Furthermore, phosphorylation of both Thr-1898 and Ser-1901 decreases the stability of the NotchICD-Maml1-CSL complex on DNA. Our data indicate that phosphorylation of Ser-1901 by CK2 leads to phosphorylation of Thr-1898 and further destabilization of the NotchICD-Maml1-CSL complex on DNA, with a concomitant decrease in transcriptional activity.
Phosphorylation within the Ankyrin Domain Destabilizes the Notch Activation Complex on DNA
In this study we demonstrate that Notch undergoes two specific phosphorylation events that negatively regulate DNA binding and transcriptional activity. The first phosphorylation event is likely to occur before NotchICD interacting with Maml1 and CSL, and the second event occurs when NotchICD is associated with Maml1 and CSL (Fig. 7; see Fig. 9). Interestingly, phosphorylation at the first site, Ser-1901, by CK2 generates a phosphorylation site at Thr-1898. This phosphorylation site does not contain a canonical consensus sequence for CK2 but can be phosphorylated by CK2 in vitro when Ser-1901 is mutated to Asp, as a new CK2 site is created (Fig. 5). Interestingly, this event led us to discover this second phosphorylation site in NotchICD. However, in vivo it is likely that a second kinase other than CK2 phosphorylates Thr-1898, as phosphorylated Ser-1901 does not conform to a canonical CK2 consensus sequence (Fig. 8B). In addition, we observe that Thr-1898 phosphorylation only happens when NotchICD forms a complex with Maml1 and CSL and after another phospho-specific event has occurred, i.e. phosphorylation of Ser-1901 by CK2. These observations suggest that these two phosphorylation events differ spatially and temporally. Previously, a component of the transcription machinery, CDK8, was shown to phosphorylate Notch within the PEST domain and negatively regulate NotchICD transcriptional activity (38, 39). Phosphorylation of Thr-1898 in cells is likely to be a nuclear-localized event as it only occurs when NotchICD forms a transcriptional activation complex. Therefore, one can hypothesize that the kinase responsible for this phosphorylation is a component of the transcription machinery, such as CDK8. Alternatively, this second kinase could be specifically recruited to NotchICD during assembly of the transcriptional machinery (Fig. 9).
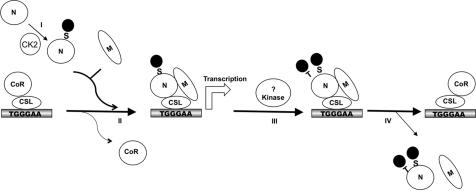
FIG. 9.
Model for hierarchical phosphorylation of NotchICD. I, NotchICD is phosphorylated by CK2 at Ser-1901 before forming an activation complex or alternatively during complex formation. II, NotchICD forms an active transcription complex with Maml1 and CSL on DNA. III, phosphorylation of Ser-1901 and interaction with Maml1 and CSL opens a second site to be phosphorylated, Thr-1898. IV, both phosphorylations will cause then the termination of transcription, which results in the displacement of NotchICD and Maml1 from DNA. CoR, corepressor.
The context of the Thr-1898 phosphorylation after the Ser-1901 phosphorylation suggests that it could be a substrate for another member of the CK family that requires phospho-priming, such as CK1. However, we tested CK1 and do not observe phosphorylation of Thr-1898 in vitro (data not shown). It is, therefore, likely that although Ser-1901 phosphorylation creates a phosphorylation site at Thr-1898, this site is exposed only when the activation complex forms and binds to DNA and might be dependent on transcription. This phosphorylation then destabilizes the active complex on DNA, resulting in the dissociation of the complex from DNA. Thus, we propose that phosphorylation of Ser-1901 followed by Thr-1898 on DNA will give a timing mechanism for transcription activation and attenuation. In this report we show that the phosphorylation events that occur at Ser-1901 and Thr-1898 are important regulators of NotchICD transcriptional activity. We have demonstrated that phosphorylation of both Ser-1901 and Thr-1898 negatively regulates NotchICD transcriptional activity by disrupting the NotchICD activation complex on DNA. How do these phosphorylation events affect the interaction between NotchICD-Maml1-CSL on DNA? One possibility is that the phosphorylation events lead to a distortion of the ankyrin domain. These phosphorylation events occur within the first ankyrin repeat of Notch and are located in a region that does not conform to a canonical ankyrin repeat. This region contains a large number of charged amino acids and is predicted to form a large flexible loop that we define now as the phosphoregulatory loop (55, 56, 62). In the crystal structure of the Notch activation complex associated with DNA, it is possible to observe that this phospho-regulatory loop is in close proximity to Maml1 and CSL, although there are no formal contacts predicted between this loop region, Maml1, and CSL (56) (Fig. 8). However, there are formal contacts between other regions of the ankyrin domain of Notch with Maml1 and CSL (55, 56). It is possible that the addition of phosphate groups in this region result in the loop undergoing a conformational change that distorts the structure of the ankyrin domain. Because both Maml1 and CSL interact with the ankyrin domain of Notch, this distortion could destabilize these interactions and affect the stability of the Notch activation complex bound to DNA. However, these phosphorylation events do not destabilize the NotchICD-Maml1-CSL complex in solution (Fig. 6E). It is possible that the Notch activation complex formed in solution is more flexible than an activation complex formed on DNA. This flexibility might allow the activation complex formed in solution to undergo more stable conformational changes, which leads to the formation of Notch activation complexes in solution containing the phosphorylated residues. Further biophysical studies need to be performed to determine how these phosphorylation events affect the structure of the Notch ankyrin domain.
Conservation of CK2 Sites in Notch Family Members
CK2 has recently been found to be involved in cell survival having an anti-apoptotic effect. Furthermore there is increasing evidence that CK2 can be responsible of tumor progression and drug resistance (63–67). In this study we showed that NotchICD is a target of CK2 and, furthermore, that this phosphorylation regulates Notch activity. This relationship raises the question if other members of the Notch family may have a similar regulation of activity by CK2. In mammals, four Notch receptors have been identified (Notch1–4) (68–71). An amino acid sequence alignment of the Notch receptors revealed that mammalian Notch1 and -2 have a conserved serine residue that conforms to a CK2 consensus sequence (Fig. 8B). Both Notch3 and Drosophila Notch do not have a serine around this position. However, they have a threonine that fits the CK2 consensus sequence, whereas Notch4 does not have either a serine or threonine in the vicinity of Ser-1901 that could be a potential CK2 site. Interestingly, Notch2 has a serine within the vicinity of Thr-1898 of Notch1, indicating that Notch2 might be regulated in a similar way as Notch1. Neither Notch3 nor Drosophila Notch has either a serine or threonine at a position similar to 1898 (Fig. 8C). Because we have described that phosphorylation of Thr-1898 occurs only in the context of complex formation on DNA, it can be predicted that Notch1 and -2 are regulated in a different manner than Notch3, Notch4, and Drosophila Notch. Moreover, Notch4, which does not have a residue equivalent to Ser-1901 or Thr-1898, may be regulated differently than any other of the Notch paralogs. The hierarchical phosphorylation that we have described in Notch1 could be a possible mechanism of differential regulation of transcription with respect to other Notch family members and opens a new field for exploration that may account for the diversity and pleiotropic effects observed in Notch signaling. Further analyses need to be performed to determine how phosphorylation of these residues contributes to the diverse biological effects observed between Notch family members.
Acknowledgments
We thank members of the Capobianco laboratory for support and technical assistance, in particular Henry Olano and Meghan Rice. We are grateful to Rhett Kovall (University of Cincinnati) for helpful discussions and critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants ROI CA 83736 and 125044-04 (NCI; to A. J. C.). This work was also supported by the Samuel Waxman Foundation for Cancer Research (to A. J. C.).
- Maml1
- Mastermind-like 1
- CK2
- casein kinase 2
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 2. Mumm J. S., Kopan R. (2000) Dev. Biol. 228, 151–165 [DOI] [PubMed] [Google Scholar]
- 3. Mumm J. S., Schroeter E. H., Saxena M. T., Griesemer A., Tian X., Pan D. J., Ray W. J., Kopan R. (2000) Mol. Cell 5, 197–206 [DOI] [PubMed] [Google Scholar]
- 4. Struhl G., Greenwald I. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vasquez-Del Carpio R., Kaplan F. M., Weaver K. L., VanWye J. D., Alves-Guerra M. C., Robbins D. J., Capobianco A. J. (2011) Mol. Cell. Biol. 31, 1396–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aster J. C., Robertson E. S., Hasserjian R. P., Turner J. R., Kieff E., Sklar J. (1997) J. Biol. Chem. 272, 11336–11343 [DOI] [PubMed] [Google Scholar]
- 7. Struhl G., Adachi A. (1998) Cell 93, 649–660 [DOI] [PubMed] [Google Scholar]
- 8. Tamura K., Taniguchi Y., Minoguchi S., Sakai T., Tun T., Furukawa T., Honjo T. (1995) Curr. Biol. 5, 1416–1423 [DOI] [PubMed] [Google Scholar]
- 9. Honjo T. (1996) Genes Cells 1, 1–9 [DOI] [PubMed] [Google Scholar]
- 10. Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. (1995) Nature 377, 355–358 [DOI] [PubMed] [Google Scholar]
- 11. Kopan R., Schroeter E. H., Weintraub H., Nye J. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1683–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schroeter E. H., Kisslinger J. A., Kopan R. (1998) Nature 393, 382–386 [DOI] [PubMed] [Google Scholar]
- 13. Weinmaster G. (1997) Mol. Cell. Neurosci. 9, 91–102 [DOI] [PubMed] [Google Scholar]
- 14. Delano W. L. (2002) The PyMOL Molecular Graphics System, Version MacPyMOL, Schrodinger, LLC, New York [Google Scholar]
- 15. Hsieh J. J., Nofziger D. E., Weinmaster G., Hayward S. D. (1997) J. Virol. 71, 1938–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohtsuka T., Ishibashi M., Gradwohl G., Nakanishi S., Guillemot F., Kageyama R. (1999) EMBO J. 18, 2196–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ronchini C., Capobianco A. J. (2001) Mol. Cell. Biol. 21, 5925–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jennings B., Preiss A., Delidakis C., Bray S. (1994) Development 120, 3537–3548 [DOI] [PubMed] [Google Scholar]
- 19. Lecourtois M., Schweisguth F. (1995) Genes Dev. 9, 2598–2608 [DOI] [PubMed] [Google Scholar]
- 20. Bailey A. M., Posakony J. W. (1995) Genes Dev. 9, 2609–2622 [DOI] [PubMed] [Google Scholar]
- 21. Bellavia D., Campese A. F., Alesse E., Vacca A., Felli M. P., Balestri A., Stoppacciaro A., Tiveron C., Tatangelo L., Giovarelli M., Gaetano C., Ruco L., Hoffman E. S., Hayday A. C., Lendahl U., Frati L., Gulino A., Screpanti I. (2000) EMBO J. 19, 3337–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Capobianco A. J., Zagouras P., Blaumueller C. M., Artavanis-Tsakonas S., Bishop J. M. (1997) Mol. Cell. Biol. 17, 6265–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ellisen L. W., Bird J., West D. C., Soreng A. L., Reynolds T. C., Smith S. D., Sklar J. (1991) Cell 66, 649–661 [DOI] [PubMed] [Google Scholar]
- 24. Gallahan D., Callahan R. (1997) Oncogene 14, 1883–1890 [DOI] [PubMed] [Google Scholar]
- 25. Gallahan D., Kozak C., Callahan R. (1987) J. Virol. 61, 218–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pear W. S., Aster J. C., Scott M. L., Hasserjian R. P., Soffer B., Sklar J., Baltimore D. (1996) J. Exp. Med. 183, 2283–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robbins J., Blondel B. J., Gallahan D., Callahan R. (1992) J. Virol. 66, 2594–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soriano J. V., Uyttendaele H., Kitajewski J., Montesano R. (2000) Int. J. Cancer 86, 652–659 [DOI] [PubMed] [Google Scholar]
- 29. Weng A. P., Ferrando A. A., Lee W., Morris J. P., 4th, Silverman L. B., Sanchez-Irizarry C., Blacklow S. C., Look A. T., Aster J. C. (2004) Science 306, 269–271 [DOI] [PubMed] [Google Scholar]
- 30. Fortini M. E. (2009) Dev. Cell 16, 633–647 [DOI] [PubMed] [Google Scholar]
- 31. Redmond L., Oh S. R., Hicks C., Weinmaster G., Ghosh A. (2000) Nat. Neurosci. 3, 30–40 [DOI] [PubMed] [Google Scholar]
- 32. Kidd S., Lieber T., Young M. W. (1998) Genes Dev. 12, 3728–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ronchini C., Capobianco A. J. (2000) Oncogene 19, 3914–3924 [DOI] [PubMed] [Google Scholar]
- 34. Shimizu K., Chiba S., Hosoya N., Kumano K., Saito T., Kurokawa M., Kanda Y., Hamada Y., Hirai H. (2000) Mol. Cell. Biol. 20, 6913–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Foltz D. R., Santiago M. C., Berechid B. E., Nye J. S. (2002) Curr. Biol. 12, 1006–1011 [DOI] [PubMed] [Google Scholar]
- 36. Foltz D. R., Nye J. S. (2001) Biochem. Biophys. Res. Commun. 286, 484–492 [DOI] [PubMed] [Google Scholar]
- 37. Fryer C. J., Lamar E., Turbachova I., Kintner C., Jones K. A. (2002) Genes Dev. 16, 1397–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fryer C. J., White J. B., Jones K. A. (2004) Mol. Cell 16, 509–520 [DOI] [PubMed] [Google Scholar]
- 39. Gupta-Rossi N., Le Bail O., Gonen H., Brou C., Logeat F., Six E., Ciechanover A., Israël A. (2001) J. Biol. Chem. 276, 34371–34378 [DOI] [PubMed] [Google Scholar]
- 40. Wu G., Lyapina S., Das I., Li J., Gurney M., Pauley A., Chui I., Deshaies R. J., Kitajewski J. (2001) Mol. Cell. Biol. 21, 7403–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oberg C., Li J., Pauley A., Wolf E., Gurney M., Lendahl U. (2001) J. Biol. Chem. 276, 35847–35853 [DOI] [PubMed] [Google Scholar]
- 42. Inglés-Esteve J., Espinosa L., Milner L. A., Caelles C., Bigas A. (2001) J. Biol. Chem. 276, 44873–44880 [DOI] [PubMed] [Google Scholar]
- 43. Espinosa L., Inglés-Esteve J., Aguilera C., Bigas A. (2003) J. Biol. Chem. 278, 32227–32235 [DOI] [PubMed] [Google Scholar]
- 44. Song J., Park S., Kim M., Shin I. (2008) FEBS Lett. 582, 1693–1699 [DOI] [PubMed] [Google Scholar]
- 45. Fernandez-Martinez J., Vela E. M., Tora-Ponsioen M., Ocaña O. H., Nieto M. A., Galceran J. (2009) J. Cell Sci. 122, 1574–1583 [DOI] [PubMed] [Google Scholar]
- 46. Ishitani T., Hirao T., Suzuki M., Isoda M., Ishitani S., Harigaya K., Kitagawa M., Matsumoto K., Itoh M. (2010) Nat. Cell Biol. 12, 278–285 [DOI] [PubMed] [Google Scholar]
- 47. Rustighi A., Tiberi L., Soldano A., Napoli M., Nuciforo P., Rosato A., Kaplan F., Capobianco A., Pece S., Di Fiore P. P., Del Sal G. (2009) Nat. Cell Biol. 11, 133–142 [DOI] [PubMed] [Google Scholar]
- 48. Jeffries S., Capobianco A. J. (2000) Mol. Cell. Biol. 20, 3928–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schägger H. (2006) Nat. Protoc. 1, 16–22 [DOI] [PubMed] [Google Scholar]
- 50. Schägger H., von Jagow G. (1987) Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 51. Kameshita I., Fujisawa H. (1989) Anal. Biochem. 183, 139–143 [DOI] [PubMed] [Google Scholar]
- 52. Luo K. X., Hurley T. R., Sefton B. M. (1991) Methods Enzymol. 201, 149–152 [DOI] [PubMed] [Google Scholar]
- 53. Jeffries S., Robbins D. J., Capobianco A. J. (2002) Mol. Cell. Biol. 22, 3927–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pagano M. A., Poletto G., Di Maira G., Cozza G., Ruzzene M., Sarno S., Bain J., Elliott M., Moro S., Zagotto G., Meggio F., Pinna L. A. (2007) Chembiochem 8, 129–139 [DOI] [PubMed] [Google Scholar]
- 55. Nam Y., Sliz P., Song L., Aster J. C., Blacklow S. C. (2006) Cell 124, 973–983 [DOI] [PubMed] [Google Scholar]
- 56. Wilson J. J., Kovall R. A. (2006) Cell 124, 985–996 [DOI] [PubMed] [Google Scholar]
- 57. Kopan R., Nye J. S., Weintraub H. (1994) Development 120, 2385–2396 [DOI] [PubMed] [Google Scholar]
- 58. Wettstein D. A., Turner D. L., Kintner C. (1997) Development 124, 693–702 [DOI] [PubMed] [Google Scholar]
- 59. Kurooka H., Kuroda K., Honjo T. (1998) Nucleic Acids Res. 26, 5448–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aster J. C., Xu L., Karnell F. G., Patriub V., Pui J. C., Pear W. S. (2000) Mol. Cell. Biol. 20, 7505–7515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhou S., Fujimuro M., Hsieh J. J., Chen L., Miyamoto A., Weinmaster G., Hayward S. D. (2000) Mol. Cell. Biol. 20, 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ehebauer M. T., Chirgadze D. Y., Hayward P., Martinez Arias A., Blundell T. L. (2005) Biochem. J. 392, 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Giusiano S., Cochet C., Filhol O., Duchemin-Pelletier E., Secq V., Bonnier P., Carcopino X., Boubli L., Birnbaum D., Garcia S., Iovanna J., Charpin C. (2011) Eur. J. Cancer 47, 792–801 [DOI] [PubMed] [Google Scholar]
- 64. Lin K. Y., Tai C., Hsu J. C., Li C. F., Fang C. L., Lai H. C., Hseu Y. C., Lin Y. F., Uen Y. H. (2011) PLoS One 6, e17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tawfic S., Yu S., Wang H., Faust R., Davis A., Ahmed K. (2001) Histol. Histopathol. 16, 573–582 [DOI] [PubMed] [Google Scholar]
- 66. Kelliher M. A., Seldin D. C., Leder P. (1996) EMBO J. 15, 5160–5166 [PMC free article] [PubMed] [Google Scholar]
- 67. Brown M. S., Diallo O. T., Hu M., Ehsanian R., Yang X., Arun P., Lu H., Korman V., Unger G., Ahmed K., Van Waes C., Chen Z. (2010) Clin. Cancer Res. 16, 2295–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lardelli M., Dahlstrand J., Lendahl U. (1994) Mech. Dev. 46, 123–136 [DOI] [PubMed] [Google Scholar]
- 69. Uyttendaele H., Marazzi G., Wu G., Yan Q., Sassoon D., Kitajewski J. (1996) Development 122, 2251–2259 [DOI] [PubMed] [Google Scholar]
- 70. Weinmaster G., Roberts V. J., Lemke G. (1991) Development 113, 199–205 [DOI] [PubMed] [Google Scholar]
- 71. Weinmaster G., Roberts V. J., Lemke G. (1992) Development 116, 931–941 [DOI] [PubMed] [Google Scholar]