FIGURE 6.
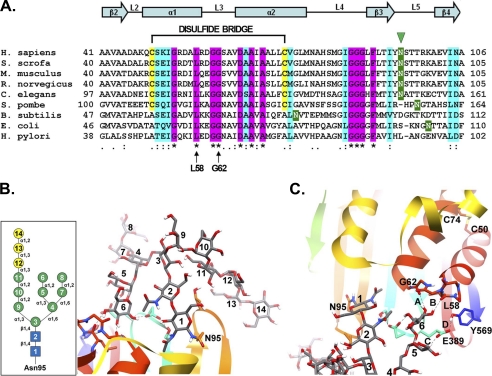
Comparative sequence and substructural analysis of the Asn-95 glycosylation site on human GGT. A, amino acid sequence alignment of residues 41–106 from human (Homo sapiens) GGT with the corresponding sequences from pig (Sus scrofa), mouse (Mus musculus), rat (Rattus norvegicus), nematode (Caenorhabditis elegans), fission yeast (Schizosaccharomyces pombe), and the bacteria Bacillus subtilis, E. coli, and Helicobacter pylori with the ClustalW program (48). Shading is used to indicate the occurrence of similar amino acids (identical residues, magenta; conserved substitutions, aquamarine) or putative N-glycosylation sites (green). Also indicated are the positions of the Asn-95 glycosylation site in human GGT (green arrowhead), the conserved glycine and leucine residues (Leu-58 and Gly-62 in human GGT) predicted to form hydrogen bonds with the N-glycan at asparagine 95 (arrows), and the C2-C3 (Cys-50 and Cys-74 in human GGT) disulfide bridge in mammalian GGT. Conserved α-helices and β-sheets are also indicated. B, homology model of human GGT propeptide showing the structural orientation of the primary oligomannosyl N-glycan (Glc3Man9GlcNAc2) conjugated to asparagine 95. Inset, structural schematic of the primary Glc3Man9GlcNAc2 carbohydrate moiety. Symbols used are as follows: blue box, N-acetylglucosamine; green circle, mannose; yellow circle, glucose. C, model of the stable hydrogen bonds (bonds A and B, dashed lines) predicted to form between a terminal mannose residue (mannose 6) in the primary N-glycan and the carbonyl oxygens of both Leu-58 and Gly-62 within the peptide loop (L3) connecting α-helices 2 and 3. Additionally, mannose 5 is predicted to hydrogen bond with Glu-389 (bond C), which is further stabilized by Tyr-569 (bond D). The position of the disulfide bond (between cysteines 50 and 74) interconnecting α-helices 2 and 3 is shown for reference.