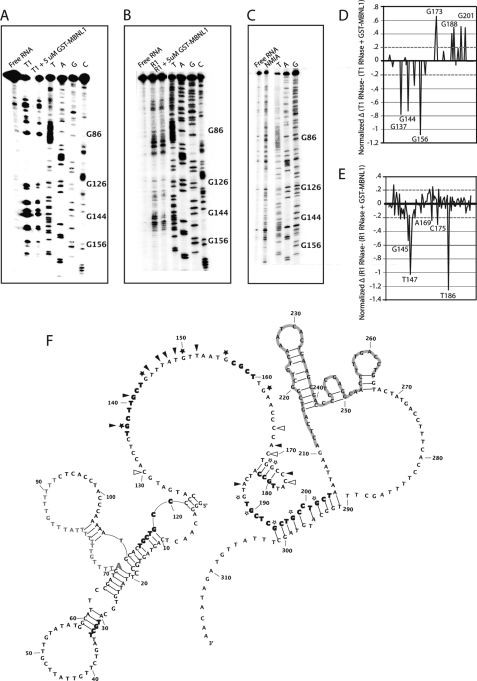
FIGURE 3.
Structure probing and footprinting of 3′-end of intron 4 of MBNL1 pre-mRNA. A, RNase T1 (cleaves single-stranded guanosines) was used for structure probing and footprinting purposes. Lane 1 is the free RNA, lane 2 has RNA and RNase T1, and lane 3 includes RNA, MBNL1, and RNase T1. B, RNase 1 (cleaves single-stranded pyrimidines) was used for structure probing and footprinting purposes. Lanes are similar to that described in A except RNase 1 is used. Lane 3 shows a footprint between nucleotides G126 and G162. C, SHAPE (used to indicate flexible nucleotides) was used to determine the secondary structure of the ultraconserved region. D, a difference plot of the RNase T1 data was created by subtracting the footprinting data (lane 3) from the structure probing data (lane 2) of the RNase T1 gels. E, a difference plot of the R1 RNase data was created in the same way as for D. The difference plots in D and E focus on the region between nucleotides G126 and G210. Nucleotides above 0.2 and below −0.2 (dashed lines shown) were considered to have increased or decreased cleavage, respectively, in the presence of MBNL1. F, experimentally derived secondary structure of the 3′-end of intron 4, exon 5, and the 5′-end of intron 5. Nucleotides 42–311 contain secondary structure data from RNase T1, RNase 1, and SHAPE gels. The structure shown is the compilation of the quantified data of the three structure probing and footprinting assays. Nucleotides that are highlighted in gray are in exon 5. The adenosine in gray with increased font size is the distant branch point, and adjacent nucleotides in gray are the PY tract. Bold nucleotides are potential MBNL1 binding site (YGCY motifs). Symbols placed next to nucleotides show changes in RNA secondary structure due to MBNL1. Black stars and arrowheads indicate nucleotides that are protected in the presence of MBNL1 in the RNase T1 and R1 RNase assays, respectively. Open stars and arrowheads indicate nucleotides that displayed enhanced cleavage in the presence of MBNL1 in the RNase T1 and RNase R1 footprinting assays.