Background: Intein activity is often dependent on the immediately flanking extein residues.
Results: Significantly improved inteins that are more promiscuous toward the flanking extein residues were generated using sequential directed evolution.
Conclusion: Sequential directed evolution is an effective tool to improve and generalize intein functionality.
Significance: Inteins being highly promiscuous toward the flanking extein residues will be important assets to improve intein-based biotechnical applications.
Keywords: Chemical Biology, Directed Evolution, Protein Chemistry, Protein Engineering, Protein Evolution, Intein, Protein Semisynthesis, Protein Splicing, Self-processing
Abstract
Inteins are internal protein sequences that post-translationally self-excise and splice together the flanking sequences, the so-called exteins. Natural and engineered inteins have been used in many practical applications. However, inteins are often inefficient or inactive when placed in a non-native host protein and may require the presence of several amino acid residues of the native exteins, which will then remain as a potential scar in the spliced protein. Thus, more general inteins that overcome these limitations are highly desirable. Here we report sequential directed evolution as a new approach to produce inteins with such properties. Random mutants of the Ssp (Synechocystis sp. PCC 6803) DnaB mini-intein were inserted into the protein conferring kanamycin resistance at a site where the parent intein was inactive for splicing. The mutants selected for splicing activity were further improved by iterating the procedure for two more cycles at different positions in the same protein. The resulting improved inteins showed high activity in the positions of the first rounds of selection, in multiple new insertion sites, and in different proteins. One of these inteins, the M86 mutant, which accumulated 8 amino acid substitutions, was also biochemically characterized in an artificially split form with a chemically synthesized N-terminal intein fragment consisting of 11 amino acids. When compared with the unevolved split intein, it exhibited an ∼60-fold increased rate in the protein trans-splicing reaction and a Kd value for the interaction of the split intein fragments improved by an order of magnitude. Implications on the intein structure-function, practical application, and evolution are discussed.
Introduction
Inteins are internal protein sequences that occur as in-frame insertions in the coding sequences of host proteins. Inteins post-translationally catalyze a protein-splicing reaction where the intein is precisely excised and the flanking sequences (N- and C-exteins) are joined by a peptide bond, producing a mature spliced protein (1). The splicing pathway, directed by the intein plus the first residue of the C-extein, typically consists of four coordinated nucleophilic displacement reactions, resulting in cleavage of the N- and C-terminal intein-extein bonds and ligation of the N- and C-exteins (2, 3). Uncoupling of these reactions, through site-directed mutations or intein placement in certain non-native host proteins, can result in N- or C-terminal cleavages without splicing. Inteins can be categorized into three types: bifunctional inteins, containing a splicing domain interrupted by a homing endonuclease domain; mini-inteins, containing only a splicing domain; and split inteins, consisting of separately encoded N- and C-inteins that can reassemble through structural complementation to catalyze trans-splicing (4, 5).
Intein-based protein splicing and cleavages have increasingly become useful tools in biological research and biotechnology. Controllable inteins have been developed as molecular switches for controlling the functions of host proteins, in which the protein-splicing function could be controlled by temperature, ligand, or light (6–10). Split inteins, capable of protein trans-splicing, have been used in transgenic plants to prevent environmental escape of the transgene (11, 12), in a gene therapy procedure to overcome size limitations of certain viral vectors (13), in two-hybrid procedures for detecting protein-protein interactions and subcellular localization (14), and for producing chemically modified (15, 16), segmentally labeled (17), and cyclic proteins, both in vitro and in vivo (18–21). The intein cleavage activities have also been developed into useful tools including the expressed protein ligation method for protein labeling/modification (22) and the Intein mediated purification with an affinity chitin-binding tag (IMPACT) method for easy purification of recombinant proteins (23).
Different inteins typically exhibit different specificities for the extein amino acid residues proximal to the intein, which can severely limit the general usefulness of intein-based methods. The intein sequence plus the first C-extein residue (invariably Ser, Cys, or Thr) generally contain all the necessary structural information for protein splicing, requiring no exogenous co-factors or energy sources. However, inteins tested outside of their native host proteins often showed inefficient splicing and/or uncoupling of the splicing reaction to yield cleavage products (24–27). The Ssp4 DnaE intein, for example, required 3 native C-extein residues for efficient trans-splicing (25). After protein splicing, these native extein residues would remain in the spliced protein, which may affect the function of the spliced protein and thus constrain where the intein can be inserted. Directed protein evolution has been used previously for engineering inteins, either by using an in vivo selection based on the reconstitution of a selectable protein through splicing (7, 28–32) or by exploiting in vitro phage display systems (33, 34). However, in these examples, either the flanking amino acids at the splice junctions have been kept constant or only a single site was used. Thus, the identification of highly promiscuous inteins that are capable of splicing in most or ideally all sequence contexts remains an important goal.
In this study, we tested whether an intein could be made more general by subjecting it to a sequential directed evolution procedure at three different insertion sites in a genetically selectable host protein. The resulting mutant inteins, when compared with the wild type intein, showed significantly improved ability of splicing at multiple new insertion sites. For one of these inteins, we also found a highly improved activity in an artificially split form that is useful for protein semisynthesis.
EXPERIMENTAL PROCEDURES
General Techniques
Unless otherwise specified, standard protocols were used. As selection markers, kanamycin (50 μg/ml) and ampicillin (100 μg/ml) were applied. Synthetic oligonucleotides were obtained from Biolegio (Nijmegen, The Netherlands) and Integrated DNA Technologies (Skokie, IL). All plasmids were verified by DNA sequencing. Reagents were purchased from Acros Organics (Nidderau, Germany), Applichem (Darmstadt, Germany), GE Healthcare (Munich, Germany), Novabiochem (Bad Soden, Germany), Roth (Karlsruhe, Germany), or Sigma-Aldrich (Munich, Germany). Restriction enzymes and markers were obtained from Fermentas (St. Leon-Rot, Germany) and New England Biolabs (Ipswich, MA). All reactions and assays were performed at least in duplicate.
Plasmid Construction
To generate the kanamycin resistance (KanR) plasmid vector, the pDrive plasmid (Qiagen) was modified by deletion of the LacZ α-peptide and multiple cloning site sequences and insertion of a His tag coding sequence at the start of the KanR gene. To facilitate intein insertion in the KanR gene, appropriate restriction sites were introduced through PCR and inverse PCR. For insertion site 1, restriction sites BspEI and SalI were inserted through silent mutations in the KanR sequence before and after the insertion site, with the corresponding amino acid sequence being -SG- and -VD-, respectively. For insertion site 2, restriction site SalI was inserted through silent mutations in the amino acid sequence -VD-, and the insertion of restriction site BglII resulted in an -SDF- to -SDL- mutation that did not prevent the KanR function. For insertion sites 3–10, restriction sites BspEI and BsrGI were inserted through silent mutations in the intein sequence, with BspEI corresponding to -SG- near the N terminus (residues 3–4) of the intein and with BsrGI corresponding to -IVH- immediately before the C terminus of the intein. Prior to directed evolution, the Ssp DnaB mini-intein coding sequence was initially amplified from pMST (35), with the appropriate restriction sites, and inserted at site 1 in the KanR plasmid vector.
The plasmid pCL20 for protein 1 (IC-Trx-His6)5 was described previously (36). For the expression plasmid pIT21 for construct 2 (ICM86-Trx-His6), the ICM86-encoding fragment was ordered as a synthetic gene from Mr. Gene (Regensburg, Germany). The DNA fragment was excised by NcoI and BamHI and ligated into a pET16b vector containing the IC-Trx-His6-encoding fragment from pCL20, in which the BamHI site between the Trx- and His6-encoding sequence was mutated to a SalI site, to give pIT21.
Directed Evolution
Error-prone PCR was performed under conditions modified from Fromant et al. (37) to amplify the intein along with ∼100 bp of flanking KanR sequence on each side of the intein. The flanking KanR sequences were included to facilitate easy purification of the intein fragment after restriction digestion of the PCR product (see below). Three sets of error-prone conditions were performed, varying by 0.25 mm Mg2+, 0.05 mm Mn2+, and 1000 μm of each dNTP in excess, 0.5 mm Mg2+, 0.1 mm Mn2+, and 1500 μm of each dNTP in excess, or 1.0 mm Mg2+, 0.2 mm Mn2+, and 2000 μm of each dNTP in excess (12 reactions in total). The PCR products were digested with a pair of restriction enzymes specified for each insertion site and separated on an agarose gel, and the extracted intein fragments were ligated into the KanR plasmid vector prepared with the same restriction enzymes. The ligation products were concentrated and purified with a Qiagen MinElute column before being used to transform Escherichia coli cells (ElectroMax cells from Invitrogen) by electroporation. The transformed cells were plated on agar plates containing 50, 75, or 150 μg/ml kanamycin for growth selection. A small aliquot of the transformed cells was plated on 50 μg/ml ampicillin to estimate the efficiency of the transformation (i.e. the size of the mutant library). Colonies grown on the kanamycin plates were collected and further grown in liquid cultures followed by analysis of protein splicing by Western blotting (Qiagen Penta-His HRP conjugate kit), determination of mutations by DNA sequencing (Macrogen Inc.) of the plasmids, or initiation of the next round of directed evolution from selected mutant intein(s).
Expression and Purification of Proteins
Protein expression and purification was performed as described previously (38). Elution fractions of the Ni2+-nitrilotriacetic acid affinity chromatography purification containing the desired protein were pooled and directly dialyzed against assay buffer (50 mm Tris, 300 mm NaCl, 1 mm EDTA, pH 7.0) with 2 mm dithiothreitol (DTT) and 10% (v/v) glycerin or anisotropy buffer (assay buffer with 2 mm tris(2-carboxyethyl)phosphine and 20% (v/v) glycerin), shock-frozen in liquid nitrogen, and stored at −80 °C.
Peptide Synthesis
Solid phase peptide synthesis of peptide 1 was performed using a Liberty peptide synthesizer (CEM, Kamp-Lintfort, Germany) on a Wang resin according to standard protocols based on the Fmoc (N-(9-fluorenyl)methoxycarbonyl) protecting group strategy and with 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate as the activation reagent. Peptide synthesis of peptides 2 and 3 as well as peptide purification and peptide analysis by MALDI-TOF MS were carried out as described previously (38).
Analysis of the Splicing Reaction by SDS-PAGE and Fluorescence Anisotropy Measurements
Splicing reaction analysis and fluorescence anisotropy measurements were performed as described previously (38).
RESULTS
Strategy for a Sequential Directed Evolution System
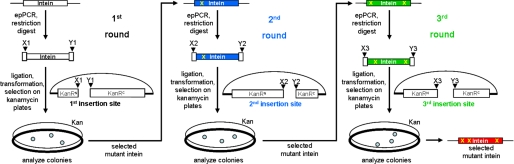
To construct a genetic selection system for the directed evolution of an intein, a KanR protein, namely aminoglycoside-3′-phospho-transferase-I (APH(3′)-I), was chosen as the host protein. After intein insertion in the KanR protein through recombinant DNA techniques, efficient protein splicing is required to restore protein function and to confer kanamycin resistance to the host E. coli cell. The KanR protein contained an N-terminal His6 tag for easy detection on Western blots. The sequential directed evolution process is illustrated in Fig. 1. The intein coding sequence was initially inserted between two endonuclease restriction sites that were introduced at a first site in the KanR-encoding gene on a plasmid vector. For directed evolution, the intein coding sequence was amplified through error-prone PCR followed by restriction digest and re-insertion of the DNA fragments into the same site by ligation. Transformation of E. coli cells with the resulting plasmids created a library of random mutants. The mutants were then subjected to genetic selection on agar plates containing kanamycin (50–150 μg/ml), allowing only functional mutants (capable of protein splicing) to grow into colonies. The selected colonies were analyzed for the expected protein splicing through Western blotting, and their plasmid DNAs were analyzed to identify mutations in the intein coding sequence. A mutant intein that spliced efficiently at the first site was evolved the same way at two other sites in KanR, where it failed to splice or showed only little activity to yield the most improved inteins after three sequential cycles.
FIGURE 1.
Schematic illustration of the sequential directed evolution. The intein coding sequence was inserted into three different sites in a plasmid-borne KanR gene. Directed evolution was performed at each site, and the selected mutations were carried over to the next site. epPCR, error-prone PCR.
Directed Evolution of a Wild Type Intein Produces the M3 Intein
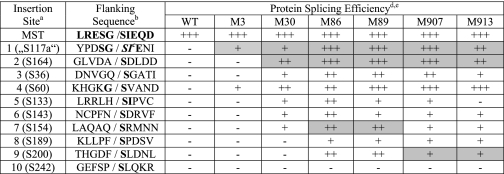
A previously reported Ssp DnaB mini-intein was chosen for this directed evolution. This intein had been known to splice efficiently (100%) in a foreign protein context (the pMST construct) when 5 native extein residues were included on each side of the intein (35). However, when this wild type intein was inserted in the KanR protein without the 5 native extein residues, the intein was unable to splice in any of the 10 insertion sites tested (Table 1). These 10 insertion sites in the KanR protein were chosen to have a Ser residue at each insertion site, which was a known requirement for splicing. The insertion sites were also chosen to be located in a variety of structural elements (supplemental Fig. S1), based on the crystal structure of a KanR homologue (Protein Data Bank no. 1ND4), the aminoglycoside-3′-phospho-transferase-IIa (APH(3′)-IIa) protein (39).
TABLE 1.
Splicing efficiencies of the Ssp DnaB wild type and improved inteins
a Sites 1–10 are insertion sites in the KanR gene.
b Native extein residues are shown in bold.
c SI are additional residues added to APH(3′)-I.
d Splicing efficiency is: −, <5%; +, 10–50%; ++, 51–80%; +++, >80%.
e Highlighted boxes indicate the insertion sites at which each improved intein was evolved through directed evolution.
The wild type Ssp DnaB mini-intein was subjected to two rounds of directed evolution at the first insertion site (site 1; Table 1) in the KanR protein. Two mutant inteins (M1 and M3) were selected with significantly improved splicing efficiencies. The M1 intein contained three mutations (K20R, D24G, and I58T) and showed a 73% splicing efficiency, which was defined as the percentage of the precursor protein that had spliced. The M3 intein showed a 36% splicing efficiency and contained two mutations (D24G and I58T).
We then tested the M3 intein for splicing at the nine other insertion sites in the KanR protein that it was not selected for. It showed significant splicing at insertion site 4 in the KanR protein, in addition to retaining the high efficiency (100%) of splicing in the original pMST construct (Table 1). However, it did not show splicing at the eight other insertion sites in the KanR protein.
Sequential Directed Evolution of the M3 Intein Produces Further Improved Mutants
The M3 intein was chosen as the starting point for further sequential rounds of directed evolution because the M1 intein was not tested at the different insertion sites in the KanR protein. A selection of mutants at the second insertion site (site 2; Table 1), where the M3 intein could not splice, yielded several new mutant inteins with a significant level of splicing, among which the M30 intein showed the highest level of splicing. Sequence determination of the M30 intein revealed four new mutations additional to those present in the M3 intein (Table 2). Unexpectedly, the amino acid residue immediately before the intein was also mutated from Ala to Gly (A[-1]G mutation). This was because the A(−1) codon was included with the intein on the SalI-BglII DNA fragment that underwent the random mutagenesis. The M30 intein showed splicing efficiencies of 98 and 71% with and without the A(−1)G mutation at insertion site 2, respectively (Table 3). The M30 intein was tested for splicing at the other insertion sites without the A(−1)G mutation. (Note: Site 4 naturally has Gly at the −1 position.) It showed significant levels (10–80%) of splicing at five other insertion sites where it had not undergone selection (Table 1). Furthermore, it retained efficient splicing in the pMST construct and at insertion site 1 in the KanR protein.
TABLE 2.
List of mutations in the improved inteins produced through directed evolution
All mutations are relative to the wild type sequence of the Ssp DnaB mini-intein.
| Mutant intein | ||||||
|---|---|---|---|---|---|---|
| M1 | M3 | M30 | M86 | M89 | M907 | M913 |
| D24G | D24G | D24G | D24G | D24G | D24G | D24G |
| I58T | I58T | I58T | I58T | I58T | I58T | I58T |
| K20R | ||||||
| S18P | S18P | S18P | S18P | S18P | ||
| S107A | S107A | S107A | S107A | S107A | ||
| S122P | S122P | S122P | S122P | S122P | ||
| H143R | H143R | H143R | H143R | H143S | ||
| S114P | ||||||
| P142L | P142L | |||||
| I30V | ||||||
| H92R | ||||||
TABLE 3.
Contributions of individual mutations to the splicing efficiency at the insertion site 2 in the KanR protein
| Mutant inteins | Mutationsa | Splicing efficiency | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| % | ||||||||||
| WT | 0 | |||||||||
| WT-1 | A(−1)G | 7 | ||||||||
| M3 | D24G | I58T | 0 | |||||||
| M3-3 | A(−)G | D24G | I58T | 65 | ||||||
| M30 | A(−)G | D24G | I58T | S18P | S107A | S122P | H143R | 98 | ||
| M30-6 | S18P | 0 | ||||||||
| M30-7 | S107A | 0 | ||||||||
| M30-8 | S122P | 0 | ||||||||
| M30-9 | H143R | 0 | ||||||||
| M30-1 | D24G | I58T | S18P | S107A | S122P | H143R | 71 | |||
| M30-2 | D24G | I58T | S18P | 22 | ||||||
| M30-3 | D24G | I58T | H143R | 38 | ||||||
| M30-4 | S18P | S107A | S122P | H143R | 15 | |||||
| M30-5 | S107A | S122P | H143R | 6 | ||||||
| M86 | D24G | I58T | S18P | S107A | S122P | H143R | S114P | P142L | 93 | |
| M86-1 | S18P | S107A | S122P | H143R | S114P | P142L | 11 | |||
| M86-2 | S107A | S122P | H143R | S114P | P142L | 20 | ||||
| M86-3 | D24G | I58T | H143R | P142L | 0 | |||||
| M86-4 | D24G | I58T | S107A | S114P | 0 | |||||
| M86-5 | D24G | I58T | S107A | 0 | ||||||
| M86-6 | D24G | I58T | S114P | 0 | ||||||
a The A(−1)G mutation is in the KanR sequence at the position immediately before the intein; all other mutations are relative to the wild type sequence of the Ssp DnaB mini-intein.
In the next round of directed evolution, random mutagenesis libraries based on the M30 intein were selected for splicing at the third insertion site (site 7; Table 1) in the KanR protein (Fig. 1). Although the M30 intein already showed a low level splicing at this site, the host E. coli cell was still sensitive to a higher concentration of kanamycin. Eight selected mutant inteins showed improved splicing, of which the two most efficient ones (M86 and M89) were chosen for further analysis. The M86 intein showed 58% splicing and contained the two new mutations S114P and P142L, whereas the M89 intein displayed 54% splicing and contained two different new mutations (I30V and H92R; Table 2). In a parallel experiment, the M30 intein was also subjected to directed evolution at another insertion site (site 9; Table 1) in the KanR protein, which was one of the three remaining sites where it was unable to splice. One round of directed evolution produced the M907 and M913 inteins, with each showing 19% splicing. The M907 intein contained one new mutation (P142L), and the M913 intein had no new mutation but altered one of the seven mutations of the M30 intein, changing the H143R mutation to the H143S mutation (Table 2).
Each of the improved inteins (M86, M89, M907, and M913) was tested for splicing in the other insertion sites in the KanR protein. They all acquired the ability to splice at insertion sites 8 and 9, although none could splice at the insertion site 10 (which has Pro at position −1, Table 1). They also retained the ability to splice at all the insertion sites where the M30 intein was able to splice, except that the M913 intein lost the ability to splice at insertion site 5 in KanR protein. Furthermore, these improved inteins showed significantly higher efficiencies of splicing at most of the insertion sites when compared with the M30 intein. The M86 intein performed the best, showing higher than 50% splicing efficiencies at most of the 10 insertion sites, with the exception of sites 8 and 10 (Table 1). These improved inteins all retained the high efficiency (nearly 100%) of splicing in the pMST construct.
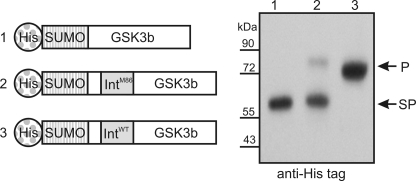
The M86 Intein Is Active in Another Host Protein
To test whether the M86 intein also performed better than the wild type Ssp DnaB mini-intein in a different host protein, the protein kinase GSK3b was chosen. The M86 intein was inserted before Ser-9 of the protein kinase, which resulted in the non-native N- and C-terminal flanking amino acids RPRTT and SFAES, respectively. The M86 intein showed efficient splicing in this new host protein, whereas the wild type Ssp DnaB mini-intein showed no detectable splicing at the same location (Fig. 2).
FIGURE 2.
Comparison of the M86 intein with the wild type intein in a protein kinase fusion protein. Left, schematic illustration of fusion protein constructs. Right, Western blot analysis of E. coli cell lysate after expression of the above fusion proteins. p = precursor protein; SP = splice product; SUMO = small ubiquitin-like modifier.
Characterization of Selected Mutations Suggests Additive Effects
We next addressed the individual contributions of the selected mutations to the activity of the improved inteins through site-directed mutagenesis. In the context of the insertion site 1 in the KanR protein, the D24G single mutation showed 11% splicing, the I58T single mutation showed 30% splicing, whereas the two mutations together (as in the M3 intein) showed 36% splicing (supplemental Table S1). The M1 intein, which had a K20R mutation additional to the D24G and I58T mutations, showed 73% splicing, although the K20R mutation alone showed only 8% splicing.
The insertion site 2 in KanR protein was used to test the effect of individual mutations of the improved inteins (Table 3). The A(−1)G mutation, which first appeared with the M30 intein, is in the KanR sequence and positioned immediately before the intein. Through site-directed mutagenesis, the A(−1)G mutation was not only found to increase the splicing efficiency of the M30 intein from 71 to 98%, but also made the wild type and the M3 inteins (which were unable to splice at this site without this mutation) splice with 7 and 65% efficiencies, respectively (Table 3). These findings suggested that the intein mutants retained a preference for the native Gly residue at the −1 position, although they had also acquired the ability to splice with different amino acids at this position. For the remaining six mutations of the M30 intein, nine different subsets (M30-1 through M30-9) were tested, and not one subset showed a splicing efficiency close to that of the M30 intein (Table 3). For the M86 intein, none of the six tested subsets of its eight mutations showed a splicing efficiency approaching that of the M86 intein having the complete set of the mutations. These results show that most, if not all, mutations are required to confer the highest splicing activity of the mutants and that these mutations contribute in an additive fashion.
Selected cis-Intein Mutations Improve trans-Splicing for Protein Semisynthesis
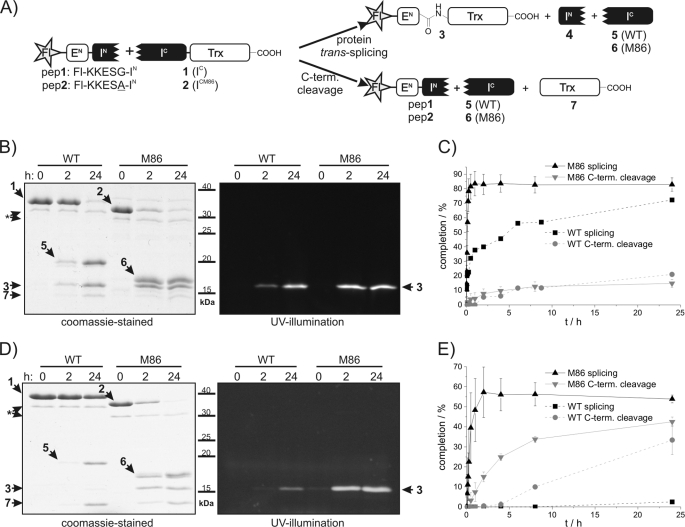
We then asked whether the selected mutations also had a beneficial effect on an artificially split variant of the Ssp DnaB intein. The split version of the wild type Ssp DnaB intein was previously shown to support protein trans-splicing when split into IN and IC fragments following amino acid 11 (S1-site) (40), and it was established as an attractive tool to ligate synthetic peptide fragments to the N terminus of proteins (36, 38). However, the preparative use of this semisynthetic split intein is limited by its relatively poor splicing kinetics and its drastically reduced activity when the native Gly(−1) is changed to another amino acid, e.g. <5% yield for Ala(−1) (38, 41). We therefore split the M86 mutant in the same way and compared the trans-splicing activity with that of the split parent intein (Fig. 3A). Note that all mutations of the M86 mutant are located in the IC fragment (amino acids 12–154). Both the original IC and the mutant ICM86 were prepared in fusion with thioredoxin (Trx) as C-extein (construct 1 = IC-Trx-His6; construct 2 = ICM86-Trx-His6) and incubated with a synthetic peptide bearing a carboxyl fluorescein moiety (Fl) as part of the N-extein (peptide 1; Fl-KKESG-IN). Fig. 3 shows that both the rate and the yield of the protein trans-splicing reaction were significantly increased for the M86 mutant intein. In agreement with previous studies (38), the parent split intein 1 approached conversion levels of ∼90% after 24 h, of which ∼75% yielded the splice product and 25% resulted in the C-terminal cleavage by-product. In comparison, the M86 split intein 2 was converted to >95% with a 5–6-fold ratio of splicing over C-terminal cleavage. The apparent first-order rate of trans-splicing was increased ∼60-fold (kWT = 4.0 ± 0.2 × 10−5 s−1, kM86 = 2.5 ± 0.1 × 10−3 s−1), such that ∼90% splice product formation was achieved already after 30 min with negligible formation of the C-terminal cleavage by-product (Fig. 3, B and C).
FIGURE 3.
Semisynthetic protein trans-splicing of the wild type and the M86 mutant inteins. A, overview of the semisynthetic protein trans-splicing reactions. pep, peptide; C-term. cleavage, C-terminal cleavage. B and D, the intein constructs 1 and 2 (15 μm) were incubated with peptides 1 and 2 (each 75 μm) for 24 h at 25 °C. Aliquots were removed at the indicated time points and analyzed by SDS-PAGE using Coomassie Brilliant Blue staining (left) and UV illumination (right). Note that construct 2 and its descendant construct 6 migrate significantly faster than constructs 1 and 5, respectively, although only differing in the eight mutations of the M86 intein. C and E, time courses of the protein trans-splicing reactions. Calculated molecular masses are: construct 1, 30.3 kDa; construct 2, 30.4 kDa; protein 3, 14.6 kDa; protein 4, 1.05 kDa; protein 5, 16.7 kDa; protein 6, 16.7 kDa; protein 7, 13.7 kDa (the asterisks denote contaminant impurity protein bands). Error bars indicate S.D.
We also studied the effect of the M86 mutations on extein sequence dependence. Consistent with previous studies (38), when wild type construct 1 was incubated with a peptide containing the Gly(−1)Ala substitution (indicated by underlining) (peptide 2; Fl-KKESA-IN), only marginal trans-splicing (∼2% after 24 h) and significant C-terminal cleavage (35% after 24 h) were observed. In contrast, construct 2 derived from the M86 mutant yielded 50% splice product and 40% C-terminal cleavage with the same peptide (Fig. 3, D and E). The rate for the trans-splicing reaction was still faster (k = 4.5 ± 0.7 × 10−4 s−1) than for the parent split intein in the context of the native Gly(−1).
Incubation of both IC constructs with a peptide containing a substitution of the active site Cys-1 to Ala (indicated by underlining) (peptide 3; Fl-KKESGAISGDSLISLA) blocked splicing, as expected. Interestingly, construct 1 was turned over to the C-terminal cleavage product in 50% yield, whereas the M86 mutant construct 2 was almost fully (96%) converted (see supplemental Fig. S2). These data showed that C-terminal cleavage is not generally suppressed in the M86 mutant, but only relative to protein trans-splicing activity.
To determine the interaction between the intein fragments, we used fluorescence anisotropy spectroscopy (38). To prevent the protein-splicing pathway following fragment association, mutations were introduced at the block B histidine (H73A) and at the C-terminal splice junction (N154A, S[+1]A) to give constructs 1a and 2a based on the wild type and the M86 mutant inteins, respectively. Consistent with the previously reported Kd for the wild type split intein (38), we determined a Kd = 0.9 ± 0.2 μm for construct 1a and peptide 1. For the M86 mutant 2a, this value was reduced by an order of magnitude (Kd = 0.10 ± 0.03 μm). A ∼2.5-fold increased kon value contributed to this improvement of the binding affinity (54.1 ± 3.4 m−1s−1 versus 20.7 ± 0.5 m−1s−1). Similarly, we measured ∼4.5-fold tighter binding and ∼2.7-fold higher kon for the M86 mutant 2a with peptide 2 containing the Gly(−1)Ala substitution (Kd = 0.5 ± 0.1 μm; kon = 60.9 ± 3.4 m−1s−1) when compared with the parent construct 1a (38).
DISCUSSION
Sequential directed evolution of the Ssp DnaB mini-intein produced several improved inteins that showed a more general ability to splice at different insertion sites in the KanR protein. This process of generating more general inteins culminated in the M86 intein, which was able to splice at 9 out of the 10 insertion sites tested in the KanR protein as well as in other host proteins. These sites confronted the intein with various different flanking residues when compared with its native sequence context. This is a dramatic improvement over the wild type intein that was unable to splice at any of these sites with non-native sequence context. Strikingly, the improved properties of the M86 intein also persisted in its artificially split form, although the selection was performed only with the cis-intein under cellular conditions, and yielded a significantly improved in vitro ligation tool for protein semisynthesis (15, 16).
The selected mutations of the improved inteins were spread over the entire intein and could not have been easily predicted by rational design (Fig. 4). Mutational analysis showed that most mutations appeared to act additively (supplemental Table S1 and Table 3). The two mutations of the M3 intein selected in the first round of the sequential evolution remained critically important for the high splicing efficiencies of the further improved inteins M30 and M86, at least when tested at the insertion site 2 in the KanR protein. The newly acquired mutations of these latter inteins significantly increased their generality in other sequence contexts. Thus, they adapted the newly improved intein to the new site of selection, but strikingly, did not lose the ability to splice at the previous sites of selection. Obviously, different beneficial mutations were accessible to genetic selection in each sequence context, and the sequential evolution approach was key to combining these mutations to act synergistically and to yield a significantly more general intein such as the M86 mutant (Table 3). This is the main difference from other studies that have reported inteins improved by directed evolution. In these cases, either the flanking residues were kept constant and close to the native sequence (7, 28, 31) or the selection was performed only at a single site in the host protein (29, 30, 32, 34). Under such conditions, it seems likely that mutant inteins evolved to a particular sequence context are selected.
FIGURE 4.
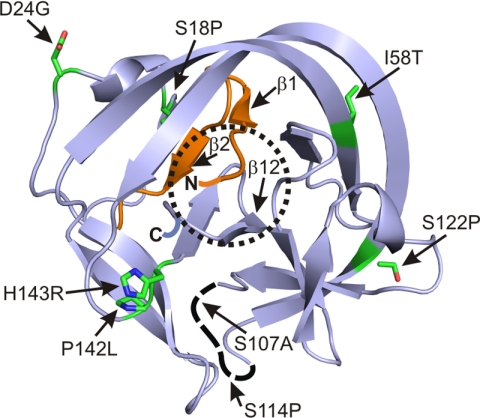
Structural presentation of the locations of mutations in the improved inteins. The ribbon structure is adapted from the crystal structure of the Ssp DnaB mini-intein (42) (Protein Data Bank no. 1MI8). The N and C termini are indicated by N and C. Mutated residues in the intein are shown as stick presentation. The active site is indicated by the dotted circle.
At the insertion site 2 in the KanR protein, the A(−1)G mutation significantly increased the splicing efficiencies of the M3 and the M30 inteins. This is not too surprising because residues other than the native Gly at this position are known to severely affect the splicing efficiency of the wild type intein (38, 41). However, the presence of Gly(−1) at the insertion sites 1 and 4 of the KanR protein was not sufficient for the wild type intein to splice. Importantly, this Gly(−1) was no longer required for the more improved inteins (e.g. the M86 intein) as these inteins spliced efficiently at other insertion sites having a variety of other amino acid residues at the −1 position (i.e. Ala, Gln, His, Asn, Phe, and Thr).
In the context of the artificially split intein useful for protein semisynthesis, the mutations of the M86 intein brought about a decrease of the Kd for the intein fragment interaction by an order of magnitude and an increase of ∼60-fold for the rate of protein trans-splicing reaction. These dramatic effects show that the mutations also improved the folding from the split fragments. The increased kon value for fragment association suggests that the mutations resulted in a better stabilized or pre-organized IC fragment.
When viewed against the crystal structure of the Ssp DnaB mini-intein, a few of the mutations are located close to the intein active site, whereas other mutations are farther away (Fig. 4). The mutations S18P and I58T are located on the opposite side of the active site on the intein surface, where they may promote the correct positioning of β-strands β1, β2, and β12 that are directly connected to the active site. The two mutations S107A and S114P are located in the loop between the IN and IC parts from which the endonuclease domain was deleted and that shows no electron density in the crystal structure of the intein (42). The H143R mutation is perhaps most interesting because His-143 is a highly conserved residue thought to be part of a charge-relay system catalyzing the Asn cyclization during splicing (42). Therefore the H143R mutation likely helped to strengthen the active site in a direct way in the improved inteins. The M913 intein, in which the H143R mutation was replaced by a H143S mutation, performed better at some sites but worse at other sites, in comparison with the M30 intein. The P142L mutation in the M86 and M907 inteins is adjacent to the H143R mutation and perhaps further strengthened the active site conformation through the H143R mutation. A mutant solely harboring the H143R mutation had no splicing activity when tested at insertion site 2 (Table 3).
In general, the inherent ability of an intein to splice is dependent upon folding into an active conformation. Achieving this conformation requires that the global intein structure is established and that the local structure at the active site is optimized for splicing. Global intein structure may be influenced by the folding dynamics of the host protein, and if the intein misfolds, it will not splice. However, even if the global intein structure is achieved, in new host contexts, suboptimal local structure at the active site of the intein might exist and result in non-splicing or cleavage instead of splicing. Our results suggest that the improved generality observed for the mutant inteins generated in this study was achieved by a greater level of structural robustness, such that the intein could possibly force its active conformation on the host protein and the directly flanking amino acids, rather than the other way around.
As the practical applications of inteins continue to expand, the availability of more general and more efficient inteins will be key to increasing the rate of success by overcoming the various degrees of extein specificities of natural and engineered inteins. Sequential directed evolution, as demonstrated in this study, provides an effective way of obtaining such improved inteins. Further rounds of sequential directed evolution might eventually lead toward a superintein that can splice in any sequence context.
Supplementary Material
Acknowledgments
We thank Simone Eppmann for technical assistance and Jens Binschik for discussion.
This work was supported by grants from the National Science and Engineering Research Council (NSERC) of Canada, the Canadian Institutes of Health Research (CIHR), the Deutsche Forschungsgemeinschaft (DFG Grant MO1073/3-1), and a Ph.D. stipend from the German National Merit Foundation (to I. V. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
IC is a wild type Ssp DnaB intein (amino acids 12–154); ICM86 is the M86 mutant of the Ssp DnaB intein (amino acids 12–154); and IN is the Ssp DnaB intein (amino acids 1–11).
- Ssp
- Synechocystis sp. PCC 6803
- Fl
- 5,6-carboxy fluorescein
- KanR
- kanamycin resistance protein
- Trx
- thioredoxin.
REFERENCES
- 1. Perler F. B., Davis E. O., Dean G. E., Gimble F. S., Jack W. E., Neff N., Noren C. J., Thorner J., Belfort M. (1994) Nucleic Acids Res. 22, 1125–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu M. Q., Perler F. B. (1996) EMBO J. 15, 5146–5153 [PMC free article] [PubMed] [Google Scholar]
- 3. Paulus H. (2000) Annu. Rev. Biochem. 69, 447–496 [DOI] [PubMed] [Google Scholar]
- 4. Liu X. Q. (2000) Annu. Rev. Genet. 34, 61–76 [DOI] [PubMed] [Google Scholar]
- 5. Saleh L., Perler F. B. (2006) Chem. Rec. 6, 183–193 [DOI] [PubMed] [Google Scholar]
- 6. Zeidler M. P., Tan C., Bellaiche Y., Cherry S., Häder S., Gayko U., Perrimon N. (2004) Nat. Biotechnol. 22, 871–876 [DOI] [PubMed] [Google Scholar]
- 7. Buskirk A. R., Ong Y. C., Gartner Z. J., Liu D. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10505–10510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mootz H. D., Blum E. S., Tyszkiewicz A. B., Muir T. W. (2003) J. Am. Chem. Soc. 125, 10561–10569 [DOI] [PubMed] [Google Scholar]
- 9. Tyszkiewicz A. B., Muir T. W. (2008) Nat. Methods 5, 303–305 [DOI] [PubMed] [Google Scholar]
- 10. Binschik J., Zettler J., Mootz H. D. (2011) Angew. Chem. Int. Ed. Engl. 50, 3249–3252 [DOI] [PubMed] [Google Scholar]
- 11. Yang J., Fox G. C., Jr., Henry-Smith T. V. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3513–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun L., Ghosh I., Paulus H., Xu M. Q. (2001) Appl. Environ. Microbiol. 67, 1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li J., Sun W., Wang B., Xiao X., Liu X. Q. (2008) Hum. Gene Ther. 19, 958–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ozawa T., Sako Y., Sato M., Kitamura T., Umezawa Y. (2003) Nat. Biotechnol. 21, 287–293 [DOI] [PubMed] [Google Scholar]
- 15. Mootz H. D. (2009) Chembiochem 10, 2579–2589 [DOI] [PubMed] [Google Scholar]
- 16. Muralidharan V., Muir T. W. (2006) Nat. Methods 3, 429–438 [DOI] [PubMed] [Google Scholar]
- 17. Busche A. E., Aranko A. S., Talebzadeh-Farooji M., Bernhard F., Dötsch V., Iwaï H. (2009) Angew. Chem. Int. Ed. Engl. 48, 6128–6131 [DOI] [PubMed] [Google Scholar]
- 18. Evans T. C., Jr., Martin D., Kolly R., Panne D., Sun L., Ghosh I., Chen L., Benner J., Liu X. Q., Xu M. Q. (2000) J. Biol. Chem. 275, 9091–9094 [DOI] [PubMed] [Google Scholar]
- 19. Iwai H., Plückthun A. (1999) FEBS Lett. 459, 166–172 [DOI] [PubMed] [Google Scholar]
- 20. Scott C. P., Abel-Santos E., Wall M., Wahnon D. C., Benkovic S. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13638–13643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Romanelli A., Shekhtman A., Cowburn D., Muir T. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6397–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muir T. W. (2003) Annu. Rev. Biochem. 72, 249–289 [DOI] [PubMed] [Google Scholar]
- 23. Chong S., Mersha F. B., Comb D. G., Scott M. E., Landry D., Vence L. M., Perler F. B., Benner J., Kucera R. B., Hirvonen C. A., Pelletier J. J., Paulus H., Xu M. Q. (1997) Gene 192, 271–281 [DOI] [PubMed] [Google Scholar]
- 24. Amitai G., Callahan B. P., Stanger M. J., Belfort G., Belfort M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11005–11010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iwai H., Züger S., Jin J., Tam P. H. (2006) FEBS Lett. 580, 1853–1858 [DOI] [PubMed] [Google Scholar]
- 26. Noren C. J., Wang J., Perler F. B. (2000) Angew. Chem. Int. Ed. Engl. 39, 450–466 [PubMed] [Google Scholar]
- 27. Sonntag T., Mootz H. D. (2011) Mol. Biosyst. 7, 2031–2039 [DOI] [PubMed] [Google Scholar]
- 28. Adam E., Perler F. B. (2002) J. Mol. Microbiol. Biotechnol. 4, 479–487 [PubMed] [Google Scholar]
- 29. Hiraga K., Derbyshire V., Dansereau J. T., Van Roey P., Belfort M. (2005) J. Mol. Biol. 354, 916–926 [DOI] [PubMed] [Google Scholar]
- 30. Lockless S. W., Muir T. W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10999–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peck S. H., Chen I., Liu D. R. (2011) Chem. Biol. 18, 619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wood D. W., Wu W., Belfort G., Derbyshire V., Belfort M. (1999) Nat. Biotechnol. 17, 889–892 [DOI] [PubMed] [Google Scholar]
- 33. Garbe D., Thiel I. V., Mootz H. D. (2010) J. Pept. Sci. 16, 575–581 [DOI] [PubMed] [Google Scholar]
- 34. Hiraga K., Soga I., Dansereau J. T., Pereira B., Derbyshire V., Du Z., Wang C., Van Roey P., Belfort G., Belfort M. (2009) J. Mol. Biol. 393, 1106–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu H., Xu M. Q., Liu X. Q. (1998) Biochim. Biophys. Acta 1387, 422–432 [DOI] [PubMed] [Google Scholar]
- 36. Ludwig C., Pfeiff M., Linne U., Mootz H. D. (2006) Angew. Chem. Int. Ed. Engl. 45, 5218–5221 [DOI] [PubMed] [Google Scholar]
- 37. Fromant M., Blanquet S., Plateau P. (1995) Anal. Biochem. 224, 347–353 [DOI] [PubMed] [Google Scholar]
- 38. Ludwig C., Schwarzer D., Mootz H. D. (2008) J. Biol. Chem. 283, 25264–25272 [DOI] [PubMed] [Google Scholar]
- 39. Nurizzo D., Shewry S. C., Perlin M. H., Brown S. A., Dholakia J. N., Fuchs R. L., Deva T., Baker E. N., Smith C. A. (2003) J. Mol. Biol. 327, 491–506 [DOI] [PubMed] [Google Scholar]
- 40. Sun W., Yang J., Liu X. Q. (2004) J. Biol. Chem. 279, 35281–35286 [DOI] [PubMed] [Google Scholar]
- 41. Mathys S., Evans T. C., Chute I. C., Wu H., Chong S., Benner J., Liu X. Q., Xu M. Q. (1999) Gene 231, 1–13 [DOI] [PubMed] [Google Scholar]
- 42. Ding Y., Xu M. Q., Ghosh I., Chen X., Ferrandon S., Lesage G., Rao Z. (2003) J. Biol. Chem. 278, 39133–39142 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.