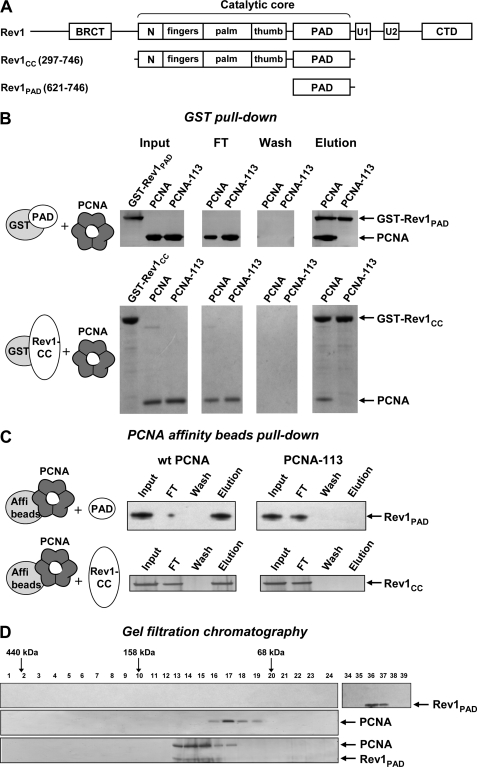
FIGURE 1.
The PAD of Rev1 interacts with PCNA at the site marked by the pol30-113 mutation. A, structure of S. cerevisiae Rev1 and its fragments used in this study. N, N-digit domain (50). U1 and U2, ubiquitin-binding motifs (23). CTD, C-terminal domain (40). B, purified GST-tagged Rev1 fragments were incubated with PCNA and glutathione-Sepharose beads and washed extensively. The bound proteins were eluted with sodium dodecyl sulfate (SDS) sample buffer, separated by electrophoresis in 4–12% SDS-polyacrylamide gel and detected by silver (top panel) or Coomassie (bottom panel) staining. FT, flow-through fraction. C, PCNA or PCNA-113 were coupled to Affi-gel 15 beads (Bio-Rad) and incubated with purified untagged Rev1 fragments. After extensive washing, the bound Rev1 fragments were eluted with SDS sample buffer, separated by electrophoresis in 4–12% SDS-polyacrylamide gel and detected by silver staining. D, Rev1-PAD (top), wild-type PCNA (middle), or the mixture of PCNA and Rev1-PAD preincubated for 1 h at 4 °C (bottom) was analyzed by gel filtration on a Superdex 200 PC 3.2/30 column (Amersham Biosciences). Fraction numbers are shown above the gel image. Ferritin (440 kDa), aldolase (158 kDa), and albumin (68 kDa) eluted at the positions indicated by the arrows. Cytochrome c (12 kDa) eluted in fractions 43–44 (not shown).