Abstract
Unlike other synthetic or physiological inhibitors for matrix metalloproteinases (MMPs), the β-amyloid precursor protein-derived inhibitory peptide (APP-IP) having an ISYGNDALMP sequence has a high selectivity toward MMP-2. Our previous study identified amino acid residues of MMP-2 essential for its selective inhibition by APP-IP and demonstrated that the N to C direction of the decapeptide inhibitor relative to the substrate-binding cleft of MMP-2 is opposite that of substrate. However, detailed interactions between the two molecules remained to be clarified. Here, we determined the crystal structure of the catalytic domain of MMP-2 in complex with APP-IP. We found that APP-IP in the complex is indeed embedded into the substrate-binding cleft of the catalytic domain in the N to C direction opposite that of substrate. With the crystal structure, it was first clarified that the aromatic side chain of Tyr3 of the inhibitor is accommodated into the S1′ pocket of the protease, and the carboxylate group of Asp6 of APP-IP coordinates bidentately to the catalytic zinc of the enzyme. The Ala7 to Pro10 and Tyr3 to Ile1 strands of the inhibitor extend into the nonprime and the prime sides of the cleft, respectively. Therefore, the decapeptide inhibitor has long range contact with the substrate-binding cleft of the protease. This mode of interaction is probably essential for the high MMP-2 selectivity of the inhibitor because MMPs share a common architecture in the vicinity of the catalytic center, but whole structures of their substrate-binding clefts have sufficient variety for the inhibitor to distinguish MMP-2 from other MMPs.
Keywords: Crystal Structure, Matrix Metalloproteinase (MMP), Peptide Interactions, Protease Inhibitor, Tumor Metastases
Introduction
The matrix metalloproteinases (MMPs)2 comprise a family of zinc-dependent endopeptidases capable of degrading protein components of the extracellular matrix and play pivotal roles in tissue remodeling under physiological and pathological conditions such as morphogenesis, angiogenesis, tissue repair, and tumor invasion (1–4). The association of MMPs with tumor invasion and metastasis has suggested that these proteases represent attractive target for the development of anti-tumor therapies. To date, a large number of MMP inhibitors based on hydroxamic acid derivatives or other synthetic inhibitors have been designed (5–8). However, none of them has been developed successfully as anti-tumor drugs mainly because of deleterious side effects; the broad specificity of the MMP inhibitors must be a stiff obstacle for developing safe and effective drugs. A common architecture of catalytic sites of MMPs probably relates to the broad specificity of the inhibitors. Moreover, recent studies (2, 9) suggest that some members of MMPs have anti-tumorigenic and anti-metastatic functions, and inhibition of their activities by broad spectrum MMP inhibitors therefore offset anti-tumor effects of the inhibitors, and even worse, stimulate tumor growth and metastasis.
The activities of MMPs in vivo are regulated by a family of inhibitors known as tissue inhibitors of metalloproteinases (TIMPs). These physiological MMP inhibitors also have broad specificity against MMPs; the activities of almost all MMPs are susceptible to TIMP (TIMP-1 to TIMP-4) inhibition, and some members of a disintegrin and a metalloproteinase family are also inhibited by these inhibitors (10, 11). In addition to the protease-inhibitory activity, some TIMPs have cell growth-stimulating activity. It has been reported recently that binding of TIMP-2 to cell surface membrane type 1-MMP (MT1-MMP) activates the ERK1/2 pathway by a nonproteolytic mechanism, thus contributing to the aggressive tumor cell migration and proliferation (12, 13). These complexities of the functions of TIMPs also make it infeasible to use the inhibitor proteins for anti-tumor therapies.
β-Amyloid precursor protein (APP) is a type I integral membrane protein, which was initially identified as a precursor of β-amyloid peptide, the principal component of extracellular deposits in senile plaques observed in Alzheimer disease brain (14). In cultured cells, APP is proteolytically cleaved at the cell surface within the β-amyloid sequence, and the extracellular domain of APP is released as a soluble APP into the culture medium (15, 16). Because the soluble APP contains an inhibitor of MMP-2 (17) and sites to interact with several components of the extracellular matrix (18–22), this secreted protein fragment is assumed to protect the extracellular matrix from the MMP-2-catalyzed degradation. Our previous study (23) demonstrated that the inhibitor is localized within the ISYGNDALMP sequence corresponding to residues 586–595 of APP770 and a synthetic decapeptide containing this sequence, named APP-derived inhibitory peptide (APP-IP), has MMP-2-selective inhibitory activity. So far, the APP-IP region is the only one physiological inhibitor that has high selectivity toward one MMP.
To clarify the mechanism of the selective inhibition, we have determined the amino acid residues of MMP-2 essential for its interaction with APP-IP by analyzing APP-IP inhibitions of various chimeric mutants of MMPs, and we have revealed that several residues of MMP-2 located far from the catalytic zinc in the nonprime or the prime side of the substrate-binding cleft are essential for the selective interaction (24). Our study also demonstrated that the N to C direction of APP-IP relative to the substrate-binding cleft of the protease is opposite that of substrate peptide. Although a novel mode of interaction between APP-IP and MMP-2 has been suggested, detailed interactions have still remained to be clarified.
Here, we describe the crystal structure of the catalytic domain of MMP-2 in complex with APP-IP and discuss how the decapeptide inhibitor can bind selectively with the active site of MMP-2. Clarification of the detailed mechanism of inhibition provides the potential to develop specific inhibitors for other individual MMPs.
EXPERIMENTAL PROCEDURES
Materials
The sources of materials used are as follows: pFLAG-CTC vector from Sigma. cDNA of human proMMP-2 was cloned into pCMV6 vector from OriGene Technologies (Rockville, MD). PrimeStar Max DNA polymerase was from Takara Bio Co. (Shiga, Japan), Affi-Gel 10 was from Bio-Rad. All custom oligo-DNA primers were provided by Rikaken Co., Ltd. (Tokyo). All other chemicals were of analytical grade or the highest quality commercially available.
Construction of Expression Vectors for the Catalytic Domain of MMP-2 and Its Mutants
The previously constructed pFLAG-N-ins-proMMP-7 vector (25) and proMMP-2-pCMV6 vector were used for the following construction. The cDNA sequence corresponding to the 7 amino acid residues in the C-terminal part of the propeptide region, the catalytic domain, and three fibronectin-like type II domains inserted in the catalytic domain of MMP-2 was amplified by PCR, using a pair of primers 5′-AAAACCGCGGAGCGGCAACCCAGATGTGG-3′ (sense primer containing the SacII site) and 5′-TTTGAATTCTTAGTCAGGAGAGGCCCCATAG-3′ (antisense primer containing the EcoRI site) and the cDNA of proMMP-2 as a template. The resultant PCR product was cleaved with SacII and EcoRI and ligated into pFLAG-N-ins-proMMP-7 cleaved also with SacII and EcoRI. The resultant pFLAG-N-ins-MMP-2-cat-FN vector was used for the following construction. To remove the cording region of the three fibronectin type II domains, PCR with PrimeStar Max DNA polymerase was carried out, using a pair of primers 5′-GGTACAGCCTGTTCCTCG-3′ (sense primer) and 5′-CTTGGCCTTCTCCCAAGG-3′ (antisense primer) and the pFLAG-N-ins-MMP-2-cat-FN as a template. The primers were designed in inverted tail-to-tail directions to amplify the cloning vector together with the part of propeptide and the catalytic domain of proMMP-2 sequence. The resultant PCR product having blunted tails was self-ligated, and the resultant pFLAG-N-ins-MMP-2-cat vector was used for the following constructions. To introduce a mutation that substitutes Glu121, the active site residue of MMP-2, with Ala, PCR with PrimeStar Max DNA polymerase was carried out, using a pair of primers 5′-GCCCACGCGTTTGGCCACGCCATGG-3′ (sense primer) and 5′-GCCAAACGCGTGGGCTGCCACGAGG-3′ (antisense primer) and the pFLAG-N-ins-MMP-2-cat as a template. The primers having a 15-base overlapped sequence including the mutagenic one were designed in inverted tail-to-tail directions to amplify the cloning vector together with the part of MMP-2 sequence described above. The resultant PCR product having adhesive tails due to the overlapped sequence was used directly for transformation according to the manufacturer's instruction. The resultant pFLAG-N-ins-MMP-2-cat (E121A) vector was used for the following construction. To introduce mutations in a loop region of the catalytic domain of MMP-2, by which crystallization of the recombinant protein is probably facilitated, PCR with PrimeStar Max DNA polymerase was carried out, using a pair of primers 5′-GGAAAAGGCGTTGGGTACAGCCTGTTCCTC-3′ (sense primer) and 5′-CCCAACGCCTTTTCCCAAGGTCCATAGCTCA-3′ (antisense primer) and the pFLAG-N-ins-MMP-2-cat (E121A) as a template. The primers having a 15-base overlapped sequence including the mutagenic one that substitute Glu108 and Gln110 of the catalytic domain of MMP-2 with Lys and Val, respectively, were designed in inverted tail-to-tail directions to amplify the cloning vector together with the part of MMP-2 sequence described above. The resultant pFLAG-N-ins-MMP-2-cat (E108K/Q110V/E121A) vector was used for expression of the recombinant protein.
Expression and Preparation of the Catalytic Domain of MMP-2 and Its Mutant
The expression vectors pFLAG-N-ins-MMP-2-cat and pFLAG-N-ins-MMP-2-cat (E108K/Q110V/E121A) were transfected separately into the Escherichia coli strain DH5α. The transformants were cultured in 2× YT medium (0.08% (w/v) tryptone, 0.5% (w/v) yeast extract, and 0.25% (w/v) NaCl) at 37 °C, and the recombinant proteins were induced by the addition of 1.0 mm isopropyl-β-d-thiogalactopyranoside. After a 5-h induction, E. coli cells were broken in 50 mm Tris-HCl (pH 8.0) containing 50 mm NaCl, 5 mm EDTA and 1% Triton X-100 with a sonicator, and the resultant inclusion bodies were collected by centrifugation. The inclusion bodies were washed three times with Tris-HCl (pH 8.0) containing 50 mm NaCl, 5 mm EDTA by sonication followed by centrifugation, and then they were solubilized in 50 mm Tris-HCl (pH 8.0) containing 6 m guanidine HCl and 100 mm dithiothreitol with gentle stirring for 2 h at 25 °C. The dissolved samples were then refolded by rapid dilution method using a refolding buffer containing 1.0 m arginine as described previously (25). The refolded proteins were dialyzed extensively against 50 mm sodium HEPES (pH 7.5) containing 150 mm NaCl and 10 mm CaCl2. The samples were clarified by centrifugation and concentrated until their concentrations became approximately 0.5 mg/ml, using a Centriprep YM-10 ultrafiltration device (Millipore Corp., Bedford, MA). The protein preparations showed >80% homogeneity as judged by SDS-PAGE. In case of the recombinant catalytic domain of MMP-2, the N-terminal 35 amino acid residues sequence preceding Tyr1, which is the N-terminal residue of the active form of MMP-2, was autocatalytically removed during the dialysis. However, the additional N-terminal sequence of the catalytically inactive mutant was not removed after dialysis, as expected. To remove the N-terminal sequence, 10 ml of the mutant catalytic domain of MMP-2 solution (0.5 mg/ml) prepared as described above was mixed with 1.0 ml of MMP-2-cat-Affi-Gel 10 in which 1.0 mg of the catalytic domain of MMP-2 was coupled with 1.0 ml of Affi-Gel 10 and incubated at 37 °C for 3 h with rotation. After incubation, the protease-coupled beads were removed by filtration, and the filtrate was further concentrated using a Centricon YM-10 ultrafiltration device (Millipore).
Co-crystallization of the Catalytic Domain of MMP-2 (E108K/Q110V/E121A) and APP-IP
One hundred microliters of MMP-2-cat (E108K/Q110V/E121A) of which the concentration was adjusted to be 15 mg/ml was mixed with 15 μl of 5 mm APP-IP to allow the two molecules to form an equimolar complex. Crystals of MMP-2-cat (E108K/Q110V/E121A) in complex with an APP-IP were obtained by hanging-drop vapor diffusion method. Rod-shaped crystals were obtained using a reservoir solution containing of 16% (w/v) PEG6000 and 0.01 m trisodium citrate at 297 K for several days. Prior to x-ray experiments, crystals were transferred to cryoprotectant consisting of the reservoir solution containing 25% (v/v) ethylene glycol. Then, the crystal was cooled in a nitrogen gas stream at 100 K. X-ray data collection was carried out using an UltraX in-house x-ray generator with an R-AXIS IV++ detector (Rigaku, Japan). X-ray diffraction data were integrated, scaled, and averaged using the program HKL2000 (26). The crystal structure of MMP-2-cat (E108K/Q110V/E121A) in complex with an APP-IP was determined by molecular replacement method with the program MOLREP (27), using the structure of MMP-2 (28) (Protein Data Base (PDB) ID code 1QIB) (28).The structure was manually improved by the program COOT (29) and refined by the programs CNS (30) and REFMAC (31). Crystallographic statistics are given in Table 1. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID code 3AYU).
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell. Ramachandran statistics indicate the fraction of residues in the most favored/allowed/disallowed regions, respectively.
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a (Å) | 61.8 |
| b (Å) | 76.0 |
| c (Å) | 37.0 |
| Resolution range (Å) | 50.0–2.00 (2.07–2.00) |
| Observed reflections | 83,976 |
| Unique reflections | 12,160 |
| Rmerge | 0.073 (0.175) |
| Completeness (%) | 97.5 (94.9) |
| <I>/σ<I> | 19.6 (14.7) |
| Refinement | |
| Resolution range (Å) | 20.0–2.00 |
| R/Rfree | 0.162/0.204 |
| Root mean square deviation bond distances (Å)/angles (°) | 0.009/1.091 |
| Ramachandran statistics (%) | 88.8/11.2/0 |
| Protein Data Bank ID code | 3AYU |
RESULTS
Primary Structure and Numbering for Amino Acid Residues of Mutant Form of the Catalytic Domain of MMP-2 Co-crystalized with APP-IP
We first tried to co-crystalize the catalytic domain of MMP-2 with APP-IP by extensive screening, but no crystal was obtained. Then, according to the previous report (28), we used a mutant form of the catalytic domain of MMP-2 for the co-crystallization and obtained a crystal of the MMP-2 mutant in complex with the peptide inhibitor. The amino acid sequence of the crystalized MMP-2 mutant, named MMP-2-cat (E108K/Q110V/E121A), which has Glu121 in the active site replaced with Ala, and residues Glu108 and Gln110 in a loop region of the catalytic domain replaced with Lys and Val, respectively, is shown in Fig. 1. To facilitate understanding, we use the catalytic domain numbering and APP-IP numbering for the amino acid residue numbers of the MMP-2 mutant and those of APP-IP, respectively. Therefore, the residues Tyr1 to Gln110 and Gly111 to Asp167 of the catalytic domain of MMP-2 correspond to residues Tyr81 to Gln190 and Gly365 to Asp421 of proMMP-2, respectively, and residues Ile1 to Pro10 of APP-IP correspond to residues Ile586 to Pro595 of APP770.
FIGURE 1.
Amino acid sequence of MMP-2-cat (E108K/Q110V/E121A). Hyphens represent the identical residues between the catalytic domain of MMP-2 (MMP-2-cat) and MMP-2-cat (E108K/Q110V/E121A) (Mutant). The boldface letters in the sequence represent the active site residues of MMPs. FN × 3 represents the site of residues deleted to excise the fibronectin-like type II repeat region of MMP-2.
Structure of APP-IP Bound to the MMP-2 Mutant
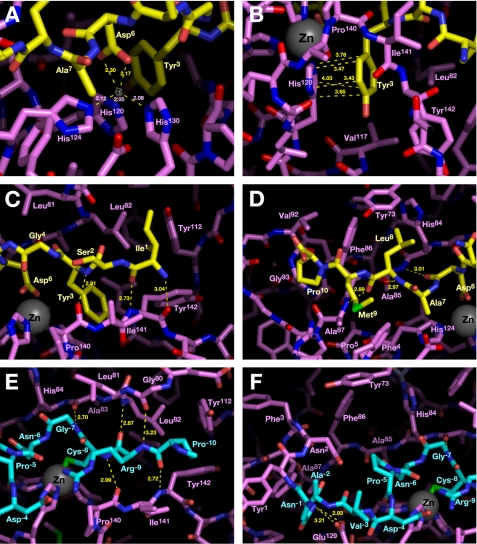
The crystal structure of MMP-2-cat (E108K/Q110V/E121A) in complex with APP-IP was determined at 2.0 Å resolution (Fig. 2A). In the crystal structure, APP-IP, the decapeptide inhibitor, is embedded in the substrate-binding cleft of the protease mutant in an almost extended conformation (Fig. 2, B and C). The N to C direction of APP-IP relative to the cleft is opposite that of substrate, as predicted previously (24). Only a part of the main chain corresponding to Gly4 and Asn5 of the inhibitor slightly loops out of the cleft, and the hydrophilic carboxamide group of Asn5 is exposed into the solvent. The residues of Tyr3 and Asp6, neighboring the small loop region of the inhibitor, are positioned in the hydrophobic subsite 1′ (S1′) pocket and close to the catalytic zinc atom of the protease, respectively (Fig. 2, B and C). The Ala7 to Pro10 and Tyr3 to Ile1 strands of the inhibitor extend into the nonprime and the prime sides of the cleft, respectively.
FIGURE 2.
Structure of the MMP-2 mutant bound to APP-IP. A, the MMP-2 mutant and APP-IP shown as magenta ribbons and yellow sticks, respectively. Zinc and calcium ions are shown as gray and white spheres, respectively. The refined electron density map (σA weighted 2Fo − Fc map, 2.1 Å resolution, contoured at 1.0σ) for APP-IP is shown as a blue cage. B, molecular surface of the MMP-2 mutant in complex with APP-IP. Various subsites in the substrate-binding cleft of MMP-2 are shown. C, stereo representation of B. The figures were prepared by using PyMOL.
Detailed Interactions between the MMP-2 Mutant and APP-IP
The distances between the catalytic zinc atom and two oxygen atoms of the carboxylate residue of Asp6 are 2.17 and 2.30 Å, respectively (Fig. 3A), indicating that the carboxylate group coordinates bidentately to the zinc atom. The phenol ring of Tyr3 of APP-IP is accommodated in the hydrophobic S1′ pocket of the protease. The most prominent interaction in this pocket is the parallel planer stacking between the phenol ring and an imidazole ring of His120 at a distance of 3.7 Å (Fig. 3B). The aliphatic side chains of Leu82 and Val117 in the S1′ pocket of the protease also contribute to the hydrophobic interaction with the aromatic side chain of the inhibitor. Other hydrophobic interactions between the aliphatic side chains of the inhibitor and the corresponding subsites of the protease schematically represented in Fig. 2B also make a large contribution; the residues Ile1, Ala7, Leu8, and Met9 of the inhibitor interact with the S2′ to S3′, S2, S3, and S4 subsites of the protease, respectively. Although there is no clear definition of S5 subsite in MMPs (32), the aliphatic residue of Pro10 of APP-IP has hydrophobic interactions with an edge of the substrate-binding cleft, probably corresponding to the S5 subsite or an exosite, via residues Phe86, Val92, and Ala87 of the protease. The main chain of the N-terminal Ile1-Ser2-Tyr3 region of APP-IP runs anti-parallel with that of the Pro140-Ile141-Tyr142 region of the MMP-2 mutant (Fig. 3C), whereas the C-terminal Asp6-Ala7-Leu8 region of the inhibitor runs parallel with the Ala85-Phe86-Ala87 region of the protease (Fig. 3D), and six hydrogen bonds between their main chains are formed.
FIGURE 3.
Detailed interactions between APP-IP and the MMP-2 mutant. A, catalytic zinc ion coordinated by the carboxylate group of Asp6 of APP-IP. B, details of the histidine-tyrosine stacking interaction in the S1′ pocket. C, hydrogen bonds formed between the backbone of Ile1-Ser2-Tyr3 region of APP-IP and that of Pro140-Ile141-Tyr142 region of MMP-2. D, hydrogen bonds formed between the backbone of Ala7-Leu8-Met9 region of APP-IP and that of Ala85-Phe86-Ala87 region of MMP-2. Leu8, Met9, and Pro10 of APP-IP involved in hydrophobic interactions with various subsites are also shown. E, hydrogen bonds formed between the backbone of Pro−10-Arg−9-Cys−8-Gly−7 region of the propeptide and that of Gly80-Leu81-Leu82-Ala83 or Pro140-Ile141-Tyr142 region of the catalytic domain in the crystal structure of proMMP-2 (PDB ID code 1GXD). The PRCGNPDVAN sequence region of the propeptide, corresponding to residues −10 through −1, is shown as cyan sticks. F, view of the C-terminal region of the propeptide corresponding to residues −6 through −1 in the structure of proMMP-2.
Based on the crystal structure of proMMP-2 (33), the C-terminal region of the propeptide, containing the PRCGNPDVAN sequence that precedes the Tyr1 of the catalytic domain of MMP-2, interacts intramolecularly with the substrate-binding cleft in the N to C direction also opposite that of substrate. The mode of interaction of the region of the propeptide with the cleft of the protease is compared with that of APP-IP in Fig. 3, E and F. The numbers −10 through −1 are given for the amino acid residue numbers of the PRCGNPDVAN sequence region. The main chain of the N-terminal Pro−10-Arg−9-Cys−8-Gly−7 region of the propeptide runs parallel and anti-parallel with that of Gly80-Leu81-Leu82-Ala83 and Pro140-Ile141-Tyr142 regions of the catalytic domain of MMP-2, respectively, and five hydrogen bonds between their main chains are formed (Fig. 3E). The thiol side chain of Cys−8 involved in the cysteine switch mechanism coordinates to the catalytic zinc ion. On the other hand, the C-terminal region of the propeptide corresponding to residues −6 through −1 has no hydrogen bond with main chain of the protease (Fig. 3F). Although the side chains of Pro−10 and Asn−6 of the propeptide interact with the S2′ and S1 subsites, respectively, no other side chain of the PRCGNPDVAN region has direct interaction with the subsites of MMP-2, indicating that the interaction between the region of the propeptide and the protease is supported mainly by the localized inter-main chain hydrogen bonds and the zinc coordination via the thiol side chain of Cys−8. In contrast, interactions of the side chains of APP-IP with their respective subsites of MMP-2 mainly stabilize the protease-inhibitor complex.
Structure of the MMP-2 Mutant Bound to APP-IP
Significant differences are apparent in the APP-IP-bound MMP-2 mutant structure compared with previously determined structure of the hydroxamate-based inhibitor-bound MMP-2 mutant (28). The two structures are superimposed with a root mean square deviation value of 0.81 Å for the comparable 160 Cα atoms (Fig. 4). The most prominent difference is found in the N-terminal region of the protease; the Tyr1 to Pro5 strand of the APP-IP-bound protease adopts ordered structure, which is stabilized partially by a salt bridge formed between the α-amino group of Tyr1 and the carboxylate group of Asp153, whereas the corresponding region of the hydroxamate inhibitor-bound one is unstructured. The conformations of the loop region corresponding to Gly107 to Gly111 are also different between the two structures, and the B-factor of this region in the hydroxamate inhibitor-bound protease is much higher than the other. Compared with the hydroxamate inhibitor-bound protease, the APP-IP-bound one has slightly wider substrate-binding cleft due to shifts in the regions corresponding to residues 73–82 and 140–143 that include the “northern” and “southern” rims of the cleft, respectively.
FIGURE 4.
Comparison of protein structures between the MMP-2 mutant bound to APP-IP and that bound to a hydroxamate inhibitor. The crystal structure of the MMP-2 mutant bound to a hydroxamate inhibitor shown in blue (PDB ID code 1QIB) and the APP-IP-bound mutant shown in magenta are superimposed. The differential regions corresponding residues 1–7, 73–82, 107–111, and 140–143 of the APP-IP-bound form are highlighted in light pink. APP-IP is shown as a stick model.
DISCUSSION
Based on the crystal structure of the MMP-2 mutant-APP-IP complex, we first identified APP-IP as a carboxylate-type metalloproteinase inhibitor in which the carboxylate group specifically coordinates with the catalytic zinc ion of the enzyme. The carboxylate side chain of Asp6 of APP-IP acts as the zinc ligand, consistent with the previous data that the substitution of the residue of the inhibitor with Ala (23) or Asn (24) leads to great loss of its inhibitory activity. To our knowledge, APP-IP is the first physiological metalloproteinase inhibitor categorized to be the carboxylate-type, although a number of synthetic carboxylate-type inhibitors have been designed. The physiological MMP inhibitors TIMPs use their N-terminal α-amino group and carbonyl oxygen of main chain of the conserved Cys1 to coordinate the catalytic zinc ion in their inhibitory actions (34–36). Compared with hydroxamate group, carboxylate group is a weaker chelator for zinc ion. The relatively weak binding of APP-IP to the catalytic center of the protease may be important for the enzyme selectivity of the inhibitor as discussed later.
APP-IP has long range contact with the substrate-binding cleft of the protease, which covers almost the entire cleft. We also found that the N to C direction of APP-IP bound to the substrate-binding cleft is opposite that of substrate peptide, consistent with our previous prediction (24). We speculate that the inversely directed interaction of APP-IP with the protease supports its inhibitory action because peptide bonds of the inhibitor are not located appropriately to the catalytic center of the enzyme, thereby making the active site-bound peptide resistant to cleavage. APP-IP is indeed resistant to MMP-2 cleavage (23). Although the propeptide of proMMP-2, an intramolecular inhibitor of the protease, also binds to the substrate-binding cleft of MMP-2 in the N to C direction opposite the substrate, its mode of interaction is significantly different from that of APP-IP; the interaction between the propeptide and the cleft is supported mainly by the localized inter-main chain hydrogen bonds and the zinc chelation by the cysteine residue in the propeptide (Fig. 3E), whereas the multiple interactions between the side chains of APP-IP and subsites of the protease mainly contribute to their high affinity interaction (Fig. 2B). Considering that the sequence of the propeptide at positions −10 through −7 is highly conserved among MMPs and the region is commonly used to inhibit the activities of MMPs in their pro-forms, the inversely directed interaction of inhibitors itself does not necessarily relate to their enzyme selectivity. The multiple side chain-subsite interactions between inhibitor and protease are thought to be rather important for the selectivity.
Our previous study (23) demonstrated that the residues Asp6 (24 kJ/mol), Leu8 (20 kJ/mol), and Tyr3 (16 kJ/mol) of APP-IP have relatively large energetic contribution in its inhibition of MMP-2 activity. In the structure of the complex, the residue Tyr3 of APP-IP is accommodated in the S1′ pocket of the protease, and its phenol group interacts with the side chain of His120 by the π-π stacking. This interaction supports the relatively high contribution of Tyr3. The π-π stacking interaction between the histidine residue of MMP-8, corresponding to His120 of MMP-2, and phenyl group of a synthetic inhibitor also has been reported (6). Many synthetic MMP inhibitors are commonly designed to have both a zinc-chelating group and a hydrophobic (aromatic or aliphatic) group accommodated in the S1′ pocket. Therefore, the YGND sequence of APP-IP containing the zinc-chelating residue Asp6 and aromatic residue Tyr3 with the linker sequence may be a minimum structural unit for MMP inhibitor. The flexibility of the linker region corresponding to Gly4 and Asn5 of APP-IP reflected by the high temperature B-factor is probably important for the appropriate positioning of the zinc-chelating and hydrophobic groups in the catalytic cleft. The high contribution of the residue Leu8 in the inhibitory activity is also supported by the hydrophobic interaction between the aliphatic residue of Leu8 and S3 subsite consisting of residues Tyr73, His84, and Phe86 of the protease. In the structure of MMP-8 inhibited by a nonprime side-directed inhibitor Pro-Leu-Gly hydroxamate, the proline residue of the inhibitor also has a hydrophobic interaction with a phenylalanine of the S3 site (37). This interaction in the S3 site explains the substrate preference of MMP-8 that cleaves peptide substrates with preference for Pro at P3 position. Considering that many MMPs as well as MMP-8 prefer Pro at P3 position of substrates (32), the hydrophobic S3 subsite is likely to be a common determinant among MMPs.
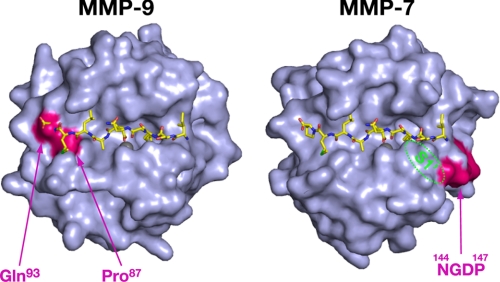
We previously identified the amino acid residues of MMP-2 essential for its selective interaction with APP-IP and found that the residues of MMP-9 at positions 87 and 93 and those of MMP-7 at positions 144–147 are the structural elements unfavorable for their interaction with APP-IP (24). When the structure of the MMP-2 mutant-APP-IP complex is superimposed with that of the catalytic domain of MMP-9 (38), the residue of Leu8 of the peptide inhibitor collides with the side chain of N-terminal Phe1 of MMP-9 because both of the residues interact with the S3 site. Considering that the structure of MMP-9 has very high B-factor in its N-terminal region corresponding to residues Phe1 to Glu2, and MMP-9 likely uses its S3 site upon peptide hydrolysis (32), the N-terminal region of the protease is assumed to move away from the substrate-binding cleft upon its interaction with peptide substrates or nonprime side-directed inhibitors. Based on this assumption, we removed the N-terminal two residues of the structure of MMP-9, and the modified structure is superimposed with that of the MMP-2 mutant-APP-IP complex (Fig. 5). We found that the C-terminal region of APP-IP still has collisional interactions with Pro87 and Gln93 of MMP-9, consistent with the previous observation that substitution of both the residue Pro87 and Gln93 of the catalytic domain of MMP-9 with Ala and Gly, respectively, enhances its affinity for the inhibitor by 196-fold (24). On the other hand, we previously speculated that Tyr3 of APP-IP locates close to Tyr144 of MMP-2 because replacement of the residues Tyr144 and Thr145 of MMP-2 with their corresponding residues of MMP-7 affects its interaction with Tyr3 of the inhibitor (24). However, the present study clarified that the aromatic side chain of the inhibitor is accommodated in the S1′ pocket and locates apart from Tyr144 of the protease. As the region of MMP-7 corresponding to residues 144–147 is located at the side of the S1′ pocket opposite where the P1′ residue of substrate or inhibitor binds (Fig. 5), it may affect the size or shape of the pocket, thereby having an adverse effect on its interaction with Tyr3 of the inhibitor.
FIGURE 5.
Structural elements in MMP-9 and MMP-7 unfavorable for their interaction with APP-IP. The crystal structure of the catalytic domain of MMP-9 (PDB ID code 1GKC) of which N-terminal two residues had been removed (see “Discussion”) or that of MMP-7 (PDB ID code 1MMQ) is superimposed with the MMP-2 mutant bound to APP-IP, and the molecular surfaces of the catalytic domain of MMP-9 (left) and MMP-7 (right) are shown in this figure. APP-IP is shown as a stick model, and the residues of MMP-9 at positions 87 and 93, and those of MMP-7 at positions 144–147 are shown in pink. The S1′ pocket located inside of MMP-7 molecule is schematically represented by a green dotted circle.
Compared with the synthetic inhibitors so far designed, APP-IP has much more groups to interact with the substrate-binding cleft of MMP-2, and some of the interactions are sensitive to the local structural variation observed among MMPs; these interactions likely contribute to the high enzyme selectivity of the inhibitor. On the other hand, the synthetic inhibitors and APP-IP share some common inhibitory mechanisms that include the chelation of the catalytic zinc ion and interaction with the hydrophobic S1′ pocket. APP-IP may be converted into a more potent inhibitor by replacing the carboxylate group of Asp6 with a hydroxamate group. Replacement of the flexible linker sequence corresponding to Gly4 and Asn5 of the inhibitor with other linkers having appropriate length and rigidity also may enhance its inhibitory activity. However, these modifications probably reduce the selectivity of the inhibitor by increasing the relative contribution of the common inhibitory mechanisms. We speculate that APP-IP uses the YGND sequence as an inhibitory unit that has the common inhibitory mechanisms with limited contribution; the minimally required contribution of the common mechanisms in the APP-IP inhibition is probably important for its selectivity.
Because some of the interactions between APP-IP and the substrate-binding cleft of MMP-2 contribute to the selectivity, substitutions of amino acid residues of APP-IP may alter its enzyme preference. As discussed above, the modifications of the residues of APP-IP other than the YGND sequence may convert it into the inhibitors that have high selectivity toward other individual MMPs. This possibility is currently under investigation in our laboratory.
This work was supported in part by 2010 Grant T2201 for Strategic Research Promotion of Yokohama City University, Japan (to S. H.) and Grants-in-aid for Scientific Research on Priority Areas 17014077 (to K. M.) and Research (C) 21570118 (to S. H.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) and the Targeted Proteins Research Program (TPRP) (to H. H. and M. S.) from MEXT.
The atomic coordinates and structure factors (code 3AYU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MMP
- matrix metalloproteinase
- APP
- β-amyloid precursor protein
- APP-IP
- APP-derived inhibitory peptide
- PDB
- Protein Data Bank
- TIMP
- tissue inhibitor of metalloproteinases.
REFERENCES
- 1. Nagase H., Visse R., Murphy G. (2006) Cardiovasc. Res. 69, 562–573 [DOI] [PubMed] [Google Scholar]
- 2. Overall C. M., Kleifeld O. (2006) Nat. Rev. Cancer 6, 227–239 [DOI] [PubMed] [Google Scholar]
- 3. Werb Z. (1997) Cell 91, 439–442 [DOI] [PubMed] [Google Scholar]
- 4. Egeblad M., Werb Z. (2002) Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 5. Betz M., Huxley P., Davies S. J., Mushtaq Y., Pieper M., Tschesche H., Bode W., Gomis-Rüth F. X. (1997) Eur. J. Biochem. 247, 356–363 [DOI] [PubMed] [Google Scholar]
- 6. Brandstetter H., Grams F., Glitz D., Lang A., Huber R., Bode W., Krell H. W., Engh R. A. (2001) J. Biol. Chem. 276, 17405–17412 [DOI] [PubMed] [Google Scholar]
- 7. Jia M. C., Schwartz M. A., Sang Q. A. (2000) Adv. Exp. Med. Biol. 476, 181–194 [DOI] [PubMed] [Google Scholar]
- 8. Finzel B. C., Baldwin E. T., Bryant G. L., Jr., Hess G. F., Wilks J. W., Trepod C. M., Mott J. E., Marshall V. P., Petzold G. L., Poorman R. A., O'Sullivan T. J., Schostarez H. J., Mitchell M. A. (1998) Protein Sci. 7, 2118–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Decock J., Thirkettle S., Wagstaff L., Edwards D. R. (2011) J. Cell Mol. Med. 15, 1254–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Amour A., Knight C. G., Webster A., Slocombe P. M., Stephens P. E., Knäuper V., Docherty A. J., Murphy G. (2000) FEBS Lett. 473, 275–279 [DOI] [PubMed] [Google Scholar]
- 11. Kashiwagi M., Tortorella M., Nagase H., Brew K. (2001) J. Biol. Chem. 276, 12501–12504 [DOI] [PubMed] [Google Scholar]
- 12. D'Alessio S., Ferrari G., Cinnante K., Scheerer W., Galloway A. C., Roses D. F., Rozanov D. V., Remacle A. G., Oh E. S., Shiryaev S. A., Strongin A. Y., Pintucci G., Mignatti P. (2008) J. Biol. Chem. 283, 87–99 [DOI] [PubMed] [Google Scholar]
- 13. Sounni N. E., Rozanov D. V., Remacle A. G., Golubkov V. S., Noel A., Strongin A. Y. (2010) Int. J. Cancer 126, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kang J., Lemaire H. G., Unterbeck A., Salbaum J. M., Masters C. L., Grzeschik K. H., Multhaup G., Beyreuther K., Müller-Hill B. (1987) Nature 325, 733–736 [DOI] [PubMed] [Google Scholar]
- 15. Sisodia S. S., Koo E. H., Beyreuther K., Unterbeck A., Price D. L. (1990) Science 248, 492–495 [DOI] [PubMed] [Google Scholar]
- 16. Esch F. S., Keim P. S., Beattie E. C., Blacher R. W., Culwell A. R., Oltersdorf T., McClure D., Ward P. J. (1990) Science 248, 1122–1124 [DOI] [PubMed] [Google Scholar]
- 17. Miyazaki K., Hasegawa M., Funahashi K., Umeda M. (1993) Nature 362, 839–841 [DOI] [PubMed] [Google Scholar]
- 18. Small D. H., Nurcombe V., Moir R., Michaelson S., Monard D., Beyreuther K., Masters C. L. (1992) J. Neurosci. 12, 4143–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cáceres J., Brandan E. (1997) J. Cell Biochem. 65, 145–158 [DOI] [PubMed] [Google Scholar]
- 20. Kibbey M. C., Jucker M., Weeks B. S., Neve R. L., Van Nostrand W. E., Kleinman H. K. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10150–10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beher D., Hesse L., Masters C. L., Multhaup G. (1996) J. Biol. Chem. 271, 1613–1620 [DOI] [PubMed] [Google Scholar]
- 22. Ohsawa I., Takamura C., Kohsaka S. (2001) J. Neurochem. 76, 1411–1420 [DOI] [PubMed] [Google Scholar]
- 23. Higashi S., Miyazaki K. (2003) J. Biol. Chem. 278, 14020–14028 [DOI] [PubMed] [Google Scholar]
- 24. Higashi S., Miyazaki K. (2008) J. Biol. Chem. 283, 10068–10078 [DOI] [PubMed] [Google Scholar]
- 25. Higashi S., Oeda M., Yamamoto K., Miyazaki K. (2008) J. Biol. Chem. 283, 35735–35744 [DOI] [PubMed] [Google Scholar]
- 26. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 28. Dhanaraj V., Williams M. G., Ye Q., Molina F., Johnson L. L., Ortwine D. F., Pavlovsky A., Rubin J. R., Skeean R. W., White A. D., Humblet C., Hupe D. J., Blundell T. L. (1999) Croat. Chem. Acta 72, 575–591 [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 31. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 32. Park H. I., Turk B. E., Gerkema F. E., Cantley L. C., Sang Q. X. (2002) J. Biol. Chem. 277, 35168–35175 [DOI] [PubMed] [Google Scholar]
- 33. Morgunova E., Tuuttila A., Bergmann U., Tryggvason K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gomis-Rüth F. X., Maskos K., Betz M., Bergner A., Huber R., Suzuki K., Yoshida N., Nagase H., Brew K., Bourenkov G. P., Bartunik H., Bode W. (1997) Nature 389, 77–81 [DOI] [PubMed] [Google Scholar]
- 35. Fernandez-Catalan C., Bode W., Huber R., Turk D., Calvete J. J., Lichte A., Tschesche H., Maskos K. (1998) EMBO J. 17, 5238–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wisniewska M., Goettig P., Maskos K., Belouski E., Winters D., Hecht R., Black R., Bode W. (2008) J. Mol. Biol. 381, 1307–1319 [DOI] [PubMed] [Google Scholar]
- 37. Bode W., Reinemer P., Huber R., Kleine T., Schnierer S., Tschesche H. (1994) EMBO J. 13, 1263–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tochowicz A., Maskos K., Huber R., Oltenfreiter R., Dive V., Yiotakis A., Zanda M., Pourmotabbed T., Bode W., Goettig P. (2007) J. Mol. Biol. 371, 989–1006 [DOI] [PubMed] [Google Scholar]