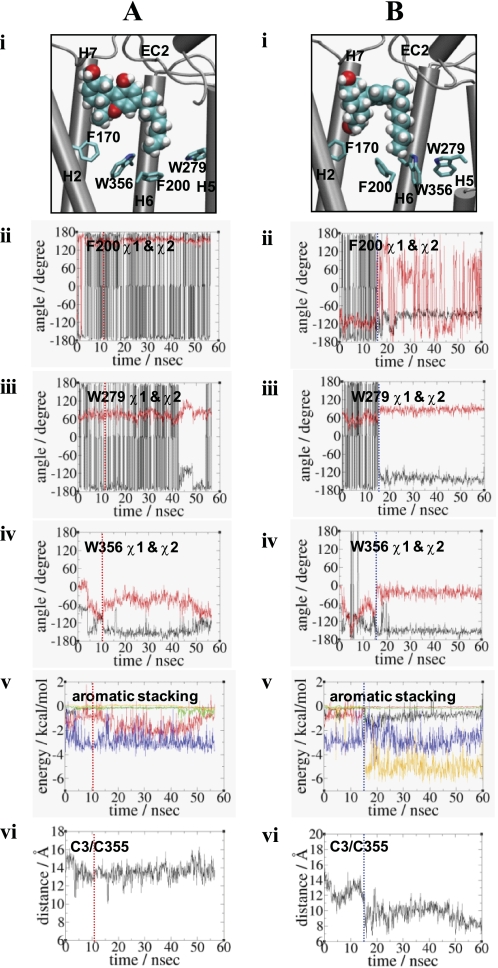
FIGURE 4.
Comparison of the Trp-3566.48 rotamer changes in HU210-CB1y2 (A) and in CP55940-CB1y (B) at the end of the simulations. Panel i, binding of the ligand (in space filling) with respect to the arrangement of Phe-1702.57, Phe-2003.36, Trp-2795.43, and Trp-3566.48 (in stick) is shown. Only the side chains without hydrogen atoms of residues are shown. H3 and H4 are omitted for a clearer view. Color coding: carbon, cyan; oxygen, red; and hydrogen, white. Panels ii–iv, rotameric angles, the χ1 angles (in black) and the χ2 angles (in red), of Phe-2003.36, Trp-2795.43, and Trp-3566.48 during the simulations. Panel v, estimated nonbonding interaction energy values between two aromatic residues, including Phe-1702.57/Phe-2003.36 (in black), Phe-1702.57/Trp-3566.48 (in red), Phe-2003.36/Trp-2795.43 (in green), Phe-2003.36/Trp-3566.48 (in blue), and Trp-2795.43/Trp-3566.48 (in orange). Only the side chains of residues were considered in calculating the nonbonding energy values. For HU210-CB1y2, the energy values (in kcal/mol), with the standard deviation in parentheses, averaged over the last 20.0 ns of the simulation are as follows: Phe-1702.57/Phe-2003.36, −0.13(0.02); Phe-1702.57/Trp-3566.48, −1.47(0.71); Phe-2003.36/Trp-2795.43, −0.33(0.24); Phe-2003.36/Trp-3566.48, −3.03(0.55); and Trp-2795.43/Trp-3566.48, −0.08(0.12). For CP55940-CB1y, the energy values (in kcal/mol), with the standard deviation in parentheses, averaged over the last 20.0 ns of the simulation are as follows: Phe-1702.57/Phe-2003.36, −0.68(0.32); Phe-1702.57/Trp-3566.48, −0.07(0.02); Phe-2003.36/Trp-2795.43, −0.17(0.04); Phe-2003.36/Trp-3566.48, −2.79(0.69); and Trp-2795.43/Trp-3566.48, −5.06(0.69). Panel vi, distance between the end carbon atom of the C3 alkyl chain of the ligand and the side chain sulfur atom of Cys-3556.47. In panels ii–vi, the time when the rotamer change occurred is represented by a dotted line: for HU210-CB1y2, in red and for CP55940-CB1y, in blue.