Fig. 4.
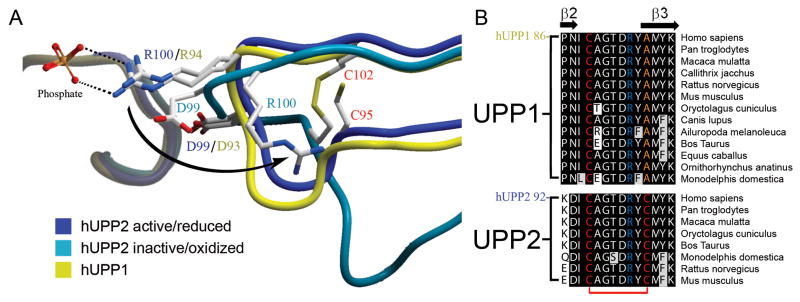
Redox inactivation of hUPP2. (A) Alternate structures of hUPP2 reveal two different conformations for the loop bearing a critical phosphate coordinating residue (R100). In one case, the structure is virtually identical to that seen in hUPP1 and other uridine phosphorylases (blue vs. yellow). However, in the second case, the formation of a disulfide bridge between C95 and C102 (red) contorts the shape of this loop and twists the key arginine residue away from the active site (turquoise). Additionally, an acidic residue (D99) is shifted closer to the phosphate binding site, which is unoccupied in this structure. These structural changes suggest that this is a catalytically incompetent conformation of this enzyme. (B) Alignment of all known mammalian UPP sequences shows that the second of the cysteine residues needed for disulfide bridge formation is invariably conserved among UPP2 proteins, but absent from all UPP1 enzymes.