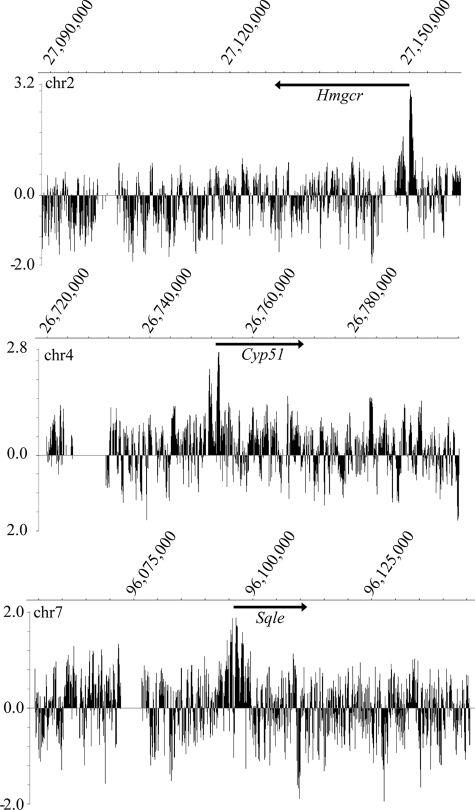
FIGURE 3.
ChIP-chip shows that Egr1 selectively binds promoters of cholesterol biosynthetic genes. The binding of Egr1 to many cholesterol biosynthetic genes was determined using ChIP-chip assays of H4IIE cells treated with insulin. The ChIP samples were labeled with Cy5 (Egr1 + insulin) or Cy3 (Egr1 − insulin control) for hybridization to the genomic tiling array. The enrichment ratio of Cy5 to Cy3 was plotted on a log2 scale and further processed to display a five-point moving average. Genomic location of peaks is displayed on the x axis. Peaks of Egr1 binding coincide with transcription start sites of cholesterol biosynthetic genes such as Hmgcr, Cyp51, and Sqle. Arrows indicate gene location and transcriptional direction. These data are representative of three independent ChIP-chip experiments, and false discovery rates values for these identified peaks are <0.05 using NimbleScan analysis.