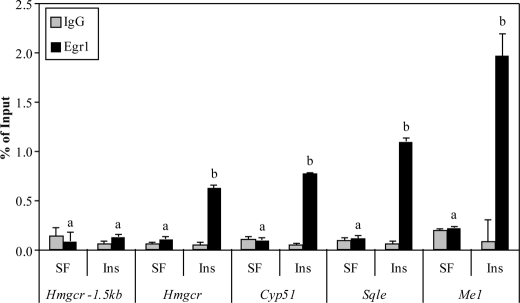
FIGURE 4.
Validation of newly identified Egr1 binding sites by ChIP. Gene elements identified as putative Egr1 binding sites were interrogated for Egr1 binding following insulin treatment for 1 h using ChIP assays. The percentage of recovery relative to input DNA was measured using qPCR with primers designed to detect putative EGR binding sites. The treatment of the cells is indicated on the x axis (SF = serum free, Ins = 1 h of insulin), as is the promoter being tested, and darker bars indicate the percentage of recovery of Egr1 at the locus, whereas lighter bars indicate the level observed with nonspecific IgG immunoprecipitation. The Hmgcr −1.5-kb primer set was used as a locus-specific negative control. Graphs represent average values from two separate experiments, data are representative of five independent experiments, and error bars indicate S.D. Bars labeled with different letters indicate statistically significant inductions of Egr1 binding (Welch's t test, p < 0.05).