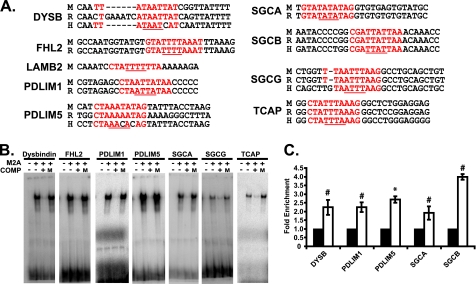
FIGURE 5.
Mef2A binds candidate Mef2 sites in costamere target gene promoters. A, conservation of Mef2 sites, from top to bottom, mouse (M), rat (R), human (H). The position of the Mef2 site relative to predicted transcription start site in the mouse gene is: DYSB (−4887), FHL2 (−1932), LAMB2 (−2767), PDLIM (−627), PDLIM5 (−304), SGCA (−497), SGCB (−3602), SGCG (−838), and TCAP (−30). Mef2 sites are highlighted in red. A/T core sequences are underlined in red. B, EMSA analysis of candidate Mef2 sites. The reactions were loaded as follows: radioactively labeled oligonucleotides with unprogrammed lysate (first lane of each gel from the left) or Mef2A protein (second lane). Unlabeled oligonucleotides containing WT (third lane) but not mutant (fourth lane) sites were able to effectively compete for binding of the labeled sites at a 50-fold molar excess. C, ChIP analysis was performed with a negative control antibody (IgG) or a Mef2A polyclonal antibody. Chromatin was subjected to quantitative PCR with primers designed to amplify ∼100 bp of promoter sequence containing the Mef2 site. Mef2A was significantly enriched on the DYSB, PDLIM1, PDLIM5, SGCA, and SGCB promoters. *, p ≤ 0.05; #, p ≤ 0.008 versus αIgG PCR values.