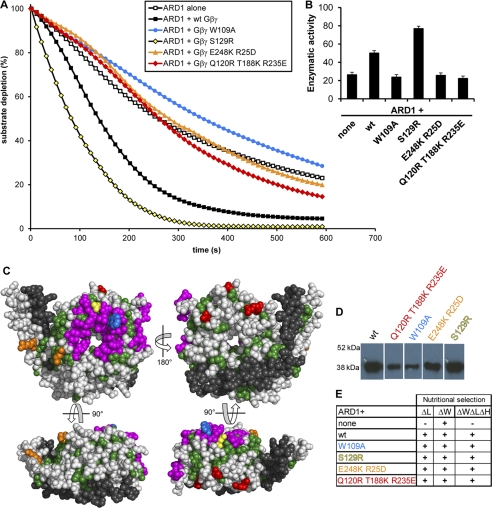
FIGURE 4.
AGB1-AGG1 stimulates ARD1, and this stimulation is reduced in several AGB1 mutants. A, ARD1 enzymatic activity in the presence and absence of wild type AGB1-AGG1 (Gβγ) and various AGB1 mutants in a 1:1 molar ratio. B, enzymatic rates of ARD1 activity in the presence of each AGB1-AGG1 wild type or mutant construct indicated (none indicates ARD1 alone). The rates are expressed in moles of substrate/mol of enzyme/s ± S.E. and were recorded as initial rates. These rates account for the average oxygen-induced decay rate (base line, see “Experimental Procedures”). C, four views of the AGB1 protein surface. All colored residues compose a region strictly conserved between plant and mammalian species. Green residues are conserved, but they have no previously known function and may be required for structural integrity. Magenta residues form the Gα binding interface. Colored residues indicate point mutations created (W109A, blue; S129R, yellow; E248K/R25D, orange; Q120R, T188K, R235E, red; colors are consistent with those in A, D, and E). Gray residues are not conserved between plants and mammals. Black residues are the Gγ protein. D, AGB1 mutant proteins were co-expressed in E. coli with AGG1 and purified via the His tag on AGG1 by affinity column chromatography (immunoblot, anti-AGB1). Proteins were separated on one gel in nonadjacent lanes. E, Y3H growth of strains containing ARD1 in the presence or absence of wild type and mutant AGB1 proteins. Cells expressing the genes encoding mutant AGB1 proteins were able to grow on media lacking histidine, indicating that the mutations do not disrupt the physical interaction between AGB1 and ARD1.