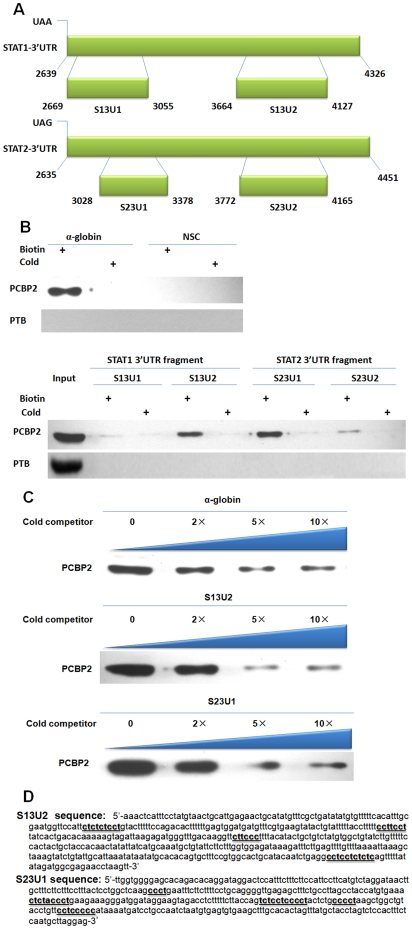
Figure 5. The C-rich tracts in the 3′UTR of mRNA of STAT1 and STAT2 are PCBP2 binding sites.
(A) Fragments of the STAT1 and STAT2 3′-UTR used in synthesis of sense RNA probe in vitro and sequences position in the 3′-UTR. (B) RNA pull-down assay using the four probes of STAT1 and STAT2. Cytoplasmic extracts from 100 IU/mL IFN-α treated R1b cells were incubated with biotin-labeled RNA probes. The mixture was pulled down by streptavidin beads. The resulting complexes were separated by SDS-PAGE and detected by Western blotting. A pGEM-3zf vector sequence was utilized as a nonspecific control (NSC), and the α-globin 3′-UTR was utilized as a positive control. The bottom panel depicts the assay results with the RNAs set of fragments shown in A. (C) 2-fold, 5-fold and 10-fold of excess unlabeled cold probes were added to compete with the labeled probes for competition analysis. The results indicate that 5-fold cold competitors of S13U2 and S23U1 probe exhibited strong competition. (D) The nucleotide sequences of S13U2 and S23U1. The CU-rich patches are underlined.