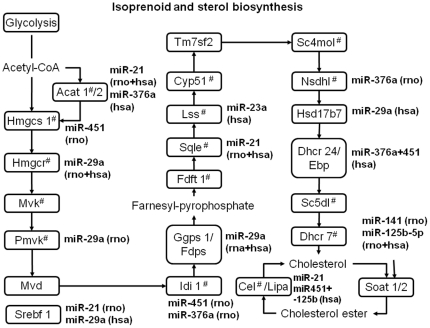
Figure 4. Enzyme pathway of isoprenoid/terpenoid and sterol biosynthesis adapted from KEGG [21] with indication of predicted target genes of regulated miRNAs in perinatal rat pancreas.
SREBF1 targets are indicated with ‘#’ next to the name and are identified through Bennett et al. (2008) and Seo et al. (2009) [48], [49]. Abbreviations: Hmgcs1 (3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble), Hmgcr (3-hydroxy-3-methylglutaryl-coA reductase), Mvk (mevalonate kinase), Pmvk (phosphomevalonate kinase), Mvd (mevalonate (diphospho) decarboxylase), Idi1 (isopentenyl-diphosphate delta isomerase 1), Fdps (farnesyl diphosphate synthase), Ggps1 (geranylgeranyl pyrophosphate synthetase), Fdft1 (farnesyl diphosphate farnesyl transferase 1), Lss (lanosterol synthase), Cyp51 (cytochrome P450, subfamily 51), Tm7sf2 (transmembrane 7 superfamily member 2), Sc4mol (sterol-C4-methyl oxidase-like), Nsdhl1 (NAD(P) dependent steroid dehydrogenase-like), Hsd17b7(hydroxysteroid (17-beta) dehydrogenase 7), Hsdl2 (hydroxysteroid dehydrogenase like 2), Dhcr24 (24-dehydrocholesterol reductase), Ebp (emopamil binding protein (sterol isomerase)), Sc5dl (sterol-C5-desaturase), Dhcr7 (7-dehydrocholesterol reductase), Soat1 (sterol O-acyltransferase 1), Cel (cholesterol ester lipase), rno (rattus norvegicus), hsa (homo sapiens).