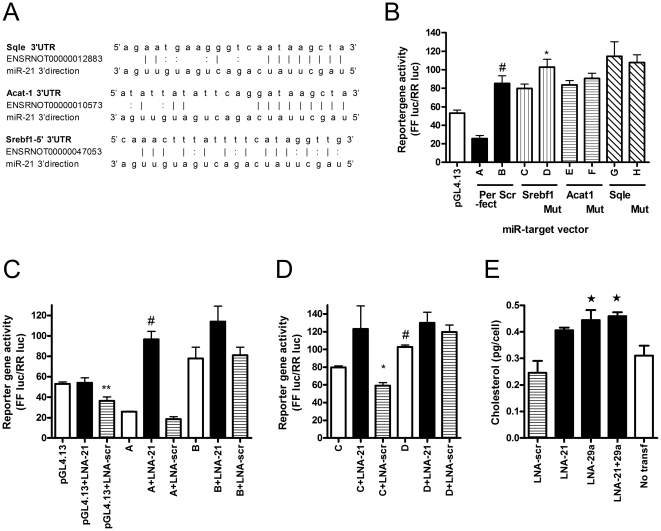
Figure 6. Reporter-gene analysis of predicted target genes of miR-21 and their binding sites.
A. Predicted binding sites in the Srebf1, Acat1 and Sqle 3′UTRs with suggested miR-21 binding. The p-values of the miRanda prediction algorithm for these three targets were: Srebf1: p = 0.0006, Acat1: p = 0.0004 and Sqle: p = 0.001. B–E. Luciferase activities of reporter vectors normalized to pRL activity. B. Activities of target site and mutant site reporter vectors together with activity of pGL4.13. (The key for clone names and their target site is: A: Perfect match for miR-21, B: Scrambled miR-21 perfect match site, C: miR-21 Srebf1 site, D: miR-21 Srebf1 site with 2 bases mutated in seed sequence, E: miR-21 Acat1 site, F: miR-21 Acat1 site with 2 bases mutated in seed sequence, G: miR-21 Sqle site with 2 bases mutated in seed sequence.) #: p<0.0001 vs. ‘A’ (‘Perfect’), *: p<0.05 vs. ‘C’ (Srebf1). C. Luciferase activities after co-transfecting LNA-21 and LNA-scr with Perfect (‘A’) and Scrambled (‘B’) vectors, **: p<0.01 vs. ‘pGL4.13+LNA-21’, #: p<0.0001 vs. ‘A+LNA-scr’. D. Luciferase activities of the Srebf1 target site (‘C’) and the 2 base mutated vector (‘D’) with co-transfection of LNA-21 and LNA-scr. #: p<0.0001 vs. ‘C’, *: p<0.05 vs. ‘C+LNA-21’. E. Total cholesterol levels of INS-1E cells following knock-down of miR-21 and/or miR-29a in INS-1E cells (LNA-21: Knock-down of miR-21, LNA-29a: Knock-down of miR-29a, LNA-scr: Scrambled control LNA, Untransf: Untransfected). *: p<0.05 vs. LNA-scr. The experiment was repeated 3 times.