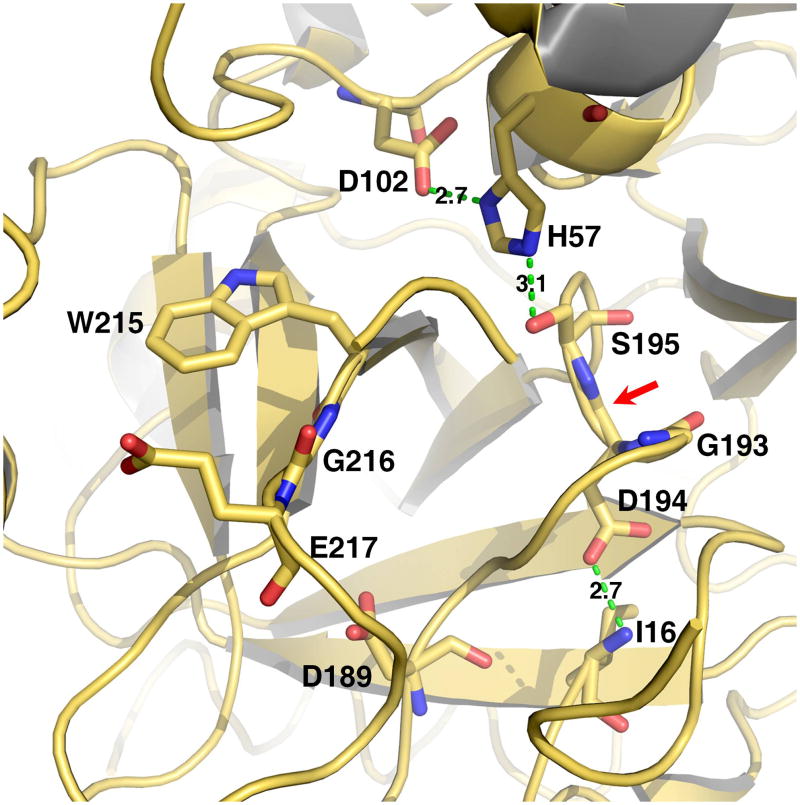
Figure 1.
Structure of the active site of a representative trypsin-like protease. Shown are the catalytic residues (H57, D102, S195), the primary specificity site (D189), the oxyanion hole formed by the backbone N atoms of S195 and G193 (red arrow), residues of the 215-217 segment and the H-bond between the N-terminus of the catalytic chain (I16) and the side chain of D194. Catalysis requires correct folding of the active site promoted by activation of the zymogen and formation of the I16-D194 H-bond, correct positioning of the catalytic residues for H transfer, correct architecture of the oxyanion hole. Binding of substrate is optimized by interaction with the primary specificity pocket (residue 189) and docking against the 215-217 segment.