Abstract
Cellular activation of latent matrix metalloproteinase-2 (proMMP-2) requires formation of a cell membrane-associated activation complex that involves specific binding between the hemopexin domain of proMMP-2 (PEX) and the C-terminal domain of tissue inhibitor of matrix metalloproteinases-2 (C-TIMP-2). In this study, we tested the feasibility of inhibiting activation of proMMP-2 by exogenous inhibitors, which block the binding between PEX and TIMP-2. The recombinant C-TIMP-2 and synthetic peptides from C-TIMP-2 were used as inhibitors for proMMP-2 activation. Recombinant C-TIMP-2 bound specifically to both the catalytically inactive MMP-2E404A and the C-terminal domain of MMP-2 (PEX) in a concentration dependent manner with apparent Kd of 3.9 × 10−7 M and 1.7 × 10−7 M, respectively. Moreover, C-TIMP-2 competed the binding between MMP-2E404A and full-length TIMP-2. Finally, activity assays showed that addition of C-TIMP-2 to HT-1080 fibrosarcoma cells inhibited proMMP-2 activation in a concentration-dependent manner. We then designed a synthetic peptide, P175L, consisting of 20 residues from the PEX-binding tail region of C-TIMP-2. P175L bound PEX and inhibited cell membrane-mediated activation of proMMP-2 in a concentration dependent manner. Deletion of the last 9 tail residues of C-TIMP-2 in P175L abrogated the inhibitory activities of the peptide showing that these residues were essential for function. Overall, these experiments have demonstrated that proMMP-2 activation can be inhibited by exogenous inhibitors which points to a potential strategy for MMP-2 specific inhibition.
Keywords: MMP-2, TIMP-2, proMMP-2 activation, peptide, MMP inhibitor, protein binding
1. Introduction
The matrix metalloproteinase family of endopeptidases (MMPs) can cleave macromolecules of the extracellular matrix and also regulates biological activities by processing and shedding bioactive molecules (Butler and Overall, 2009). In addition to their roles in normal tissue remodeling, elevated MMP activities and excessive tissue destruction have been associated with such pathological processes as tumor expansion and metastasis (Rosenthal and Matrisian, 2006), chronic inflammatory conditions (Giannobile, 2008) and periodontal disease (Sorsa et al., 2006), and abnormal wound healing in diabetes (Armstrong and Jude, 2002; Stanley et al., 2008). In an effort to inhibit MMPs in these diseases, significant effort has been devoted to develop MMP inhibitors, resulting primarily in inhibitors that were designed to mimic substrate cleavage sites and to chelate the MMP active site zinc ion. Although these compounds had good inhibitors properties, they were also characterized by low specificities due to the structural similarities in the active sites across the MMPs leading to extensive side effects in clinical trials (Brown, 1998). Consequently, investigators are seeking novel design approaches to enhance the specificities of MMP inhibitors by targeting exodomains of MMPs (Overall and Lopez-Otin, 2002). TIMPs are family of endogenous MMP inhibitors with at least four members, TIMP-1, −2, −3, and −4 (Gomez et al., 1997). All TIMPs share common structural features including an ~125 amino acid N-terminal domain and a smaller ~65-residue C-terminal domain, that are both stabilized by three disulfide bonds (Williamson et al., 1990). The N-terminal domain has inhibitory activities by binding the active sites of MMPs (Murphy et al., 1991). The C-terminal regions are divergent, which may enhance the selectivity of inhibition and binding efficiencies (Willenbrock et al., 1993).
MMPs are synthesized in latent form (proMMPs) and proteolytic removal of the prodomain is required for enzyme activation. However, MMP-2 is unique among the 25 MMPs in that it is subject to cell-mediated activation by a pathway involving the tissue inhibitor of matrix metalloproteinases-2 (TIMP-2) (Murphy et al., 1999; Strongin et al., 1993; Strongin et al., 1995). During the activation, membrane type 1-MMP (MT1-MMP) and TIMP-2 form a binary complex on cell surfaces, which in turn binds MMP-2 via the carboxyl-terminal hemopexin-like domain (PEX). After formation of this tri-molecular complex, a free MT1-MMP cleaves the pro-domain of proMMP-2 to activate the enzyme (Strongin et al., 1995). The fact that this mode of cell-mediated activation is unique to MMP-2 and that the interactions between the carboxyl-terminal domains of TIMP-2 (C-TIMP-2) and the PEX are critical for activation of proMMP-2 (Worley et al., 2003) suggest that these interactions provide a promising target for highly specific inhibition of MMP-2.
Structural studies of proMMP-2 in complex with TIMP-2 mapped how the two proteins interact via their C-terminal domains (Morgunova et al., 2002) (Fig 1D). Substitution of putative binding residues by alanine-scanning mutagenesis identified cationic, but non-contiguous residues at the junction of modules III and IV of PEX that were critical for binding of TIMP-2 (Overall et al., 1999). Reduction of the disulfide bridges and fragmentation of PEX by CNBr resulted in loss of binding to TIMP-2 implying that the tertiary structure of PEX likewise is important (Strongin et al., 1993). In comparison, the PEX binding on TIMP-2 involves a contiguous 10 amino acid C-terminal tail sequence, which is critical for the MMP-2 activation (Butler et al., 1998; Worley et al., 2003).
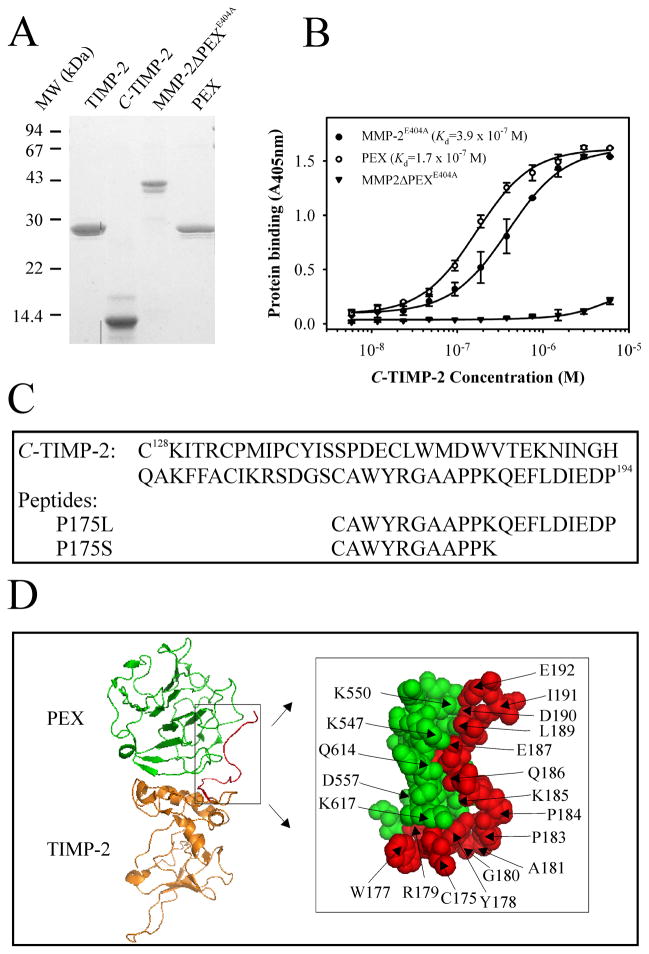
Figure 1. SDS-PAGE of recombinant proteins and Interaction of PEX with C-TIMP-2.
Panel A. Samples (~2 μg/lane) of TIMP-2, C-TIMP-2, PEX and MMP-2ΔPEXE404A were reduced with DTT, separated on 12.6 % (w/v) cross-linked polyacrylamide mini-slab gels, and stained with Coomassie Brilliant Blue R-250. The bands showed protein purity and confirmed the predicted molecular masses for each recombinant protein. The positions of protein standards (MW in kDa) are indicated. Panel B. The binding of C-TIMP-2 to MMP-2E404A, PEX, and MMP-2ΔPEXE404A was analyzed in 96 microwell plate assays. A concentration range of biotinylated C-TIMP-2 was interacted with immobilized recombinant proteins, detected by alkaline phosphatase-conjugated streptavidin with PNPP substrate and quantified at 405 mM. C-TIMP-2 bound to MMP-2E404A and PEX in a concentration-dependent manner but did not bind MMP-2ΔPEXE404A. Experiments were performed in duplicate and repeated. Data points shown are the means and S.D. (bars). Panel C. Amino acid sequence of C-terminal domain of TIMP-2 and synthetic peptides P175L and P175S are shown by one-letter codes. Panel D. Interaction between TIMP-2 and PEX is illustrated based on the coordinates for the TIPM-2/MMP-2 complex (RCSB protein data bank ID: 1GXD). PEX is in green and TIMP-2 is in orange with C- terminal tail in red. The amplified box depicts the contact region between PEX and the TIMP-2 tail labeled with one-letter codes for amino acids.
Prior studies have shown that low concentrations of exogenous of TIMP-2 stimulated activation of proMMP-2, whereas high concentrations of exogenous TIMP-2 inhibited activation of proMMP-2 (Overall et al., 2000; Strongin et al., 1993). It was also found that a recombinant C-TIMP-2 – myoglobin fusion protein could inhibit activation of proMMP-2 in fibroblast cells (Kai et al., 2002). Our present experiments confirmed those observations and tested the hypothesis that synthetic peptides derived from the tail of C-TIMP-2 could block cellular activation of MMP-2 by competing the binding between TIMP-2 and proMMP-2 based on the apparent importance of the contiguous tail residues of the C-TIMP-2 for proMMP-2 activation. The results of our experiments demonstrated that recombinant C-TIMP-2 bound specifically to PEX, competed interactions of PEX with C-TIMP-2, and inhibited cellular activation of MMP-2. Moreover, a 20-amino acid linear synthetic peptide from the tail of TIMP-2 bound specifically to PEX and inhibited MMP-2 activation by a mechanism requiring the last 9 residues of C-TIMP-2 that are critical for the interactions between TIMP-2 and PEX of MMP-2.
2. Results
2.1. Recombinant C- terminal domain of TIMP-2 (C-TIMP-2) interacts with PEX of MMP-2
The expressed and purified recombinant MMP-2ΔPEXE404A, PEX, TIMP-2 and C-TIMP-2 had the predicted masses and identities as verified by SDS-PAGE and MALDI-TOF mass spectrometry (Fig. 1A). Complementing results from prior yeast two-hybrid analysis (Overall et al., 2000), that pointed to interactions of the C-terminal domain of TIMP-2 with PEX of proMMP-2, we here confirmed that recombinant C-TIMP-2 binds both MMP-2 and the PEX domain of MMP-2 in vitro. The binding occurred in a concentration dependent manner with apparent Kds of 3.9 × 10−7 M and 1.7 × 10−7 M for MMP-2E404A and PEX, respectively (Fig. 1B). These affinities are consistent with those reported for binding between PEX and full-length TIMP-2 supporting the functional role of C-TIMP-2 in this interaction (Bigg et al., 1997; Olson et al., 1997). Importantly, C-TIMP-2 did not bind MMP-2ΔPEXE404A demonstrating that the PEX domain is responsible for the interaction with the C-terminal domain of TIMP-2.
2.2. C-TIMP-2 inhibits cell surface activation of proMMP-2
Addition of exogenous full length TIMP-2 can inhibit activation of proMMP-2 by a mechanism that has been proposed to involve competition of the binding between TIMP-2 and proMMP-2 in the activation complex (Strongin et al., 1995). However, TIMP-2 also can block activation of proMMP-2 by inhibiting the activity of MT1-MMP through interaction of the active site of the enzyme with the N-terminus of TIMP-2 (Butler et al., 1998). In the present studies, we analyzed the effects of isolated carboxyl-terminal domain of TIMP-2 (C-TIMP-2) in proMMP-2 activation assays. By this approach, we were able to exclude the effects of interactions between the N- terminal domain of TIMP-2 and MT1-MMP and to specifically test the role of C-TIMP-2 on the cellular activation of proMMP-2. To measure the effects of C-TIMP-2, we used HT1080 fibrosarcoma cells stimulated with Con A, which induces cell surface activation of proMMP-2 (Overall et al., 2000). The C-TIMP-2 inhibited activation of proMMP-2 as reflected by reduced conversion of latent to active MMP-2 in conditioned media (Fig. 2A). When quantified by densitometric analysis of the proMMP-2 band intensities on the zymography gels, the inhibitory effects of C-TIMP-2 on proMMP-2 activation were concentration dependent. At 7 μM C-TIMP-2, there was a 4-fold reduction in activation of proMMP-2 (Fig. 2B). We confirmed these results with rat osteosarcoma cells (ROS) and human head and neck squamous cell carcinoma cells (SCC-25) (not shown). It should be noted that the intensity of proMMP-2 in the conditioned medium without Con A was 39,162 RDU (Fig 2A, Lane 0 nM, Con A −). In the presence of Con A, the intensity of activated MMP-2 was 34,199 RDU and non-activated MMP-2 was 3,439 RDU (Fig 2A, Lane 0 nM, Con A +). The fact that >90% proMMP-2 could be activated by Con A confirmed that Con A is a very strong activator of proMMP-2 in cell culture. C-TIMP-2 at 7 μM inhibited about one third of the activation of proMMP-2 (13,289 RDU) (Fig 2A and B, lane 7000 nM, Con A +) under this experimental condition. However, this inhibitory efficiency of C-TIMP-2 on proMMP-2 activation may not reflect the inhibitory capacity of C-TIMP-2 under physiologic conditions.
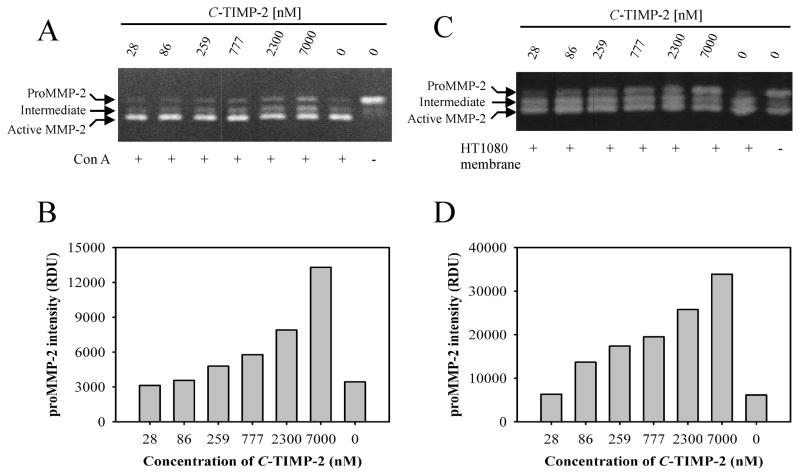
Figure 2. C-TIMP-2 inhibits cell surface activation of proMMP-2.
Panel A. Inhibition of proMMP-2 activation by C-TIMP-2 in HT1080 fibrosarcoma cell cultures. Confluent cells in 96 microwell plates in serum-free DMEM were incubated with 20 μg/ml Con A and a concentration range of C-TIMP-2 (28 – 7000 nM, 0 nM). Conditioned media were collected and the conversion of proMMP-2 to intermediate and fully activated MMP-2 was determined using 8% gelatin zymography gels. Almost all proMMP-2 was activated in the presence of Con A (C-TIMP-2; 0 nM). Addition of C-TIMP-2 to the cell culture media inhibited activation of proMMP-2 in a concentration dependent manner. Results from one representative experiment shown here were confirmed in repeated experiments. Panel B. Quantification of proMMP-2 activation from the zymography gel presented in Panel A by densitometric analysis of the proMMP-2 bands. Presented are proMMP-2 band intensities in the presence of different concentration of C-TIMP-2. Panel C. Activation of purified proMMP-2 by HT1080 cell membrane preparations. HT1080 cell membranes were isolated and incubated with proMMP-2 in the presence of a concentration range of C-TIMP (28 – 7000 nM, 0 nM). The reactions were analyzed by zymography after 16 h. C-TIMP-2 inhibited the activation of proMMP-2 by HT1080 cell membranes in a concentration dependent manner as indicated by the increasing intensity of the proMMP-2 band. Results were confirmed in repeated experiments. One representative experiment is shown. Panel D. Densitometric quantification of proMMP-2 bands from the zymography gel presented in Panel C showing band intensities in the presence of different concentrations of C-TIMP-2.
To further control the experimental conditions and reduce the variability inherent to cell culture experiments, we isolated HT1080 cell membranes as proMMP-2 activator. Preliminary experiments verified that the membrane preparations containing 1 μg total protein had enough proMMP-2 activator to effectively activate exogenous purified proMMP-2 as detected by zymography (not shown). Of note, at this amount, no membrane-bound MMP-2 was detectable. To confirm the inhibitory effects of C-TIMP-2 on cell membrane-mediated activation of proMMP-2, we incubated proMMP-2 with HT1080 membranes in the presence of a concentration range of C-TIMP-2. Zymography assays confirmed that the C-TIMP-2 inhibited activation of proMMP-2 in a concentration dependent manner in the cell-free system (Fig. 2C, D). With increasing concentrations of C-TIMP-2, we detected more latent MMP-2 and less intermediate activated MMP-2 suggesting that C-TIMP-2 inhibited the initial cleavage of proMMP-2. In contrast to the effects observed in Con A stimulated cell cultures, C-TIMP-2 at 7 μM inhibited almost all cell membrane induced proMMP-2 activation reflected by the intensity of the proMMP-2 band in the presence of C-TIMP-2 of 33,850 RDU (Fig 2C, lane 7000 nM, HT1080 membrane +) and the intensity of proMMP-2 without cell membrane activator (30715 RDU) (Fig 2C, lane 0 nM, HT1080 membrane −). Together, these results showed that interactions between PEX and the C-terminal domain of TIMP-2 in the cellular activation complex are essential for activation of proMMP-2, but not likely involving any signal transduction.
2.3. C-TIMP-2 competes the binding between TIMP-2 and MMP-2E404A
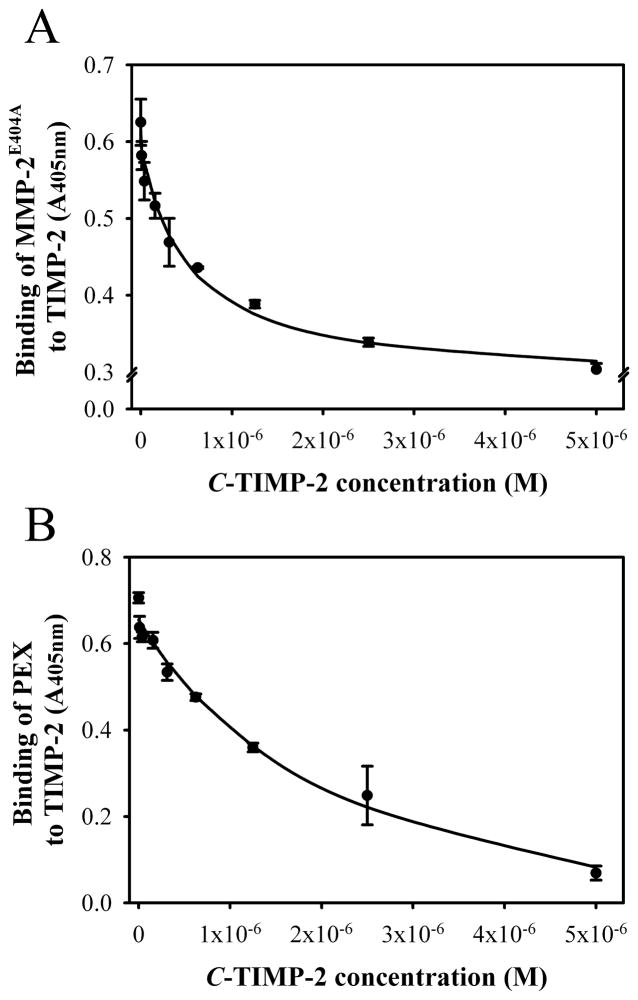
Having demonstrated that the C-TIMP-2 binds specifically to PEX of MMP-2 and inhibits the cellular activation of proMMP-2, we investigated the capacity of C-TIMP-2 to disrupt the interactions between MMP-2E404A and TIMP-2 in competitive protein-protein binding assays. In these experiments, C-TIMP-2 inhibited the binding of recombinant MMP-2E404A to TIMP-2 in a concentration-dependent manner (Fig. 3A), with a 50% of maximum inhibition being reached with 5 μM C-TIMP-2. The observation that C-TIMP-2 did not completely inhibit the binding between MMP-2E404A and TIMP-2 might be explained by the additional interactions between the catalytic domain of MMP-2 and the N-terminus of TIMP-2 (Howard and Banda, 1991). To test this possibility, we used isolated PEX domain of MMP-2 in competitive binding assays since PEX does not bind the N-terminus of TIMP-2. In those experiments, a concentration of 5 μM C-TIMP-2 inhibited the binding between PEX and TIMP-2 by ~90% (Fig. 3B). We concluded from these results that the C-terminal domain of TIMP-2 can compete the interactions of proMMP-2 and TIMP-2 and, in turn, inhibit formation of the MT1-MMP/TIMP-2/ProMMP-2 activation complex.
Figure 3. C-TIMP-2 compete the binding between TIMP-2 and MMP-2E404A.
Competitive protein – protein binding assays were carried out in 96 microwell plates. Biotinylated MMP-2E404A (Panel A) or biotinylated PEX (Panel B) were added alone or with a concentration range of unlabeled C-TIMP-2 (5000 – 10 nM) to plates coated with TIMP-2. The binding of MMP-2E404A or PEX to immobilized TIMP-2 in the presence of competitive C-TIMP-2 were detected by alkaline phosphatase-conjugated streptavidin with PNPP substrate and quantified at 405 nM. C-TIMP-2 partly inhibited binding between MMP-2E404A and TIMP-2 (Panel A) and effectively blocked PEX interaction with TIMP-2 (Panel B). Data points represent means and S.D. (bars) for two independent experiments performed in duplicate.
2.4. PEX binds C-terminal tail peptide of TIMP-2 (P175L)
Subsequent experiments explored the potential for identifying a peptide with the capacity to inhibit activation of proMMP-2. For this purpose, we designed and synthesized two TIMP-2-derived peptides based on the reported crystal structure of proMMP-2 in complex with TIMP-2 (Morgunova et al., 2002) and results from site-specific mutagenesis and deletion of putative binding site residues on TIMP-2 (Worley et al., 2003) (Fig 1D).
Although these studies predicted that the last 10-amino acid residues in the C-terminal tail of TIMP-2 were critical for PEX binding (Worley et al., 2003), neither a synthetic TIMP-2 tail peptide containing the last 13-residues nor additional peptides designed by others bound PEX or inhibited activation of proMMP-2 (Overall et al., 2000). To address these challenges, we synthesized a 20-residue C-TIMP-2 tail peptide corresponding to residues 175–194 (P175L). A shorter 11-residue control peptide, P175S, consisted C-TIMP-2 residues 175–185 also present in P175L, but lacked the last 9 C-terminal residues of TIMP-2 (See also Discussion). This experimental design enabled us to extend the analyses of the structurally identified TIMP-2 binding motif and to test the functional requirement for the last 9-residues of TIMP-2 on the binding between TIMP-2 and proMMP-2 (Worley et al., 2003).
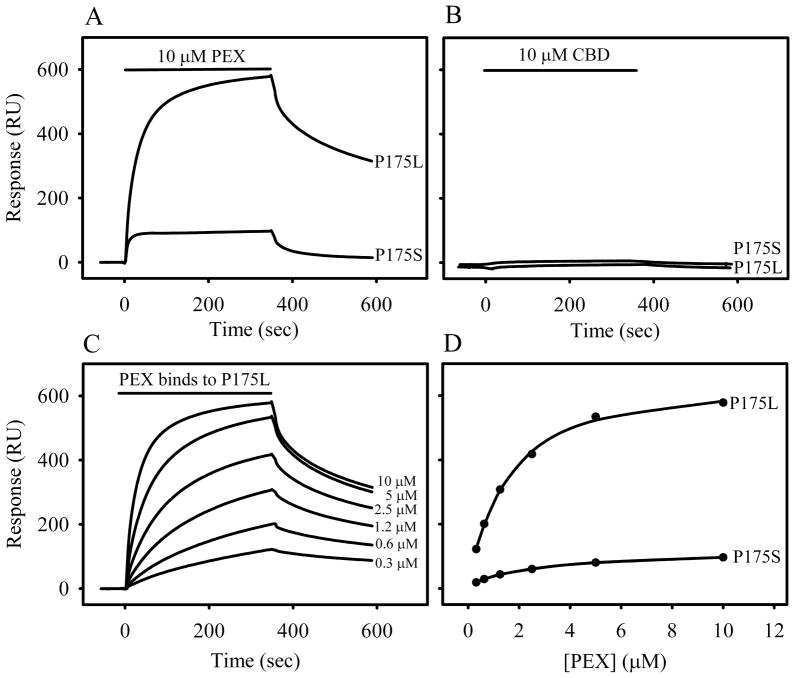
In surface plasmon resonance assays, PEX (10 μM) interacted specifically with the immobilized P175L and P175S C-TIMP-2 peptides (Fig. 4A). However, the interaction of PEX with P175L (579 RU) was about six fold higher than that of P175S (97 RUs). To confirm the specificity of the interactions between PEX and the TIMP-2 tail peptides, 10 μM recombinant collagen binding domain of MMP-2 (CBD) flowed over the same chips did not bind P175L or P175S peptides (Fig. 4B). In further experiments to characterize the interactions between PEX and the peptides, we used a range of concentrations of PEX and found that the binding between PEX and TIMP-2 peptide P175L was concentration-dependent with an apparent Kd of 1.6 × 10−6 M (Fig. 4C, D). The binding of PEX to P175S was consistently very weak. Overall, these binding assays demonstrated that P175L had sufficient length to cover the TIMP-2 binding site for PEX and confirmed previous reports indicating that the last 10 residues in the tail region of TIMP-2 are essential for the interactions (Worley et al., 2003).
Figure 4. Interaction of synthetic C-terminal tail peptides of TIMP-2 with PEX.
In SPR binding assays, synthetic peptides P175L or P175S were immobilized on CM5 chip at 327 and 319 response units, respectively. Subsequently, 30 μl aliquots of purified recombinant PEX or CBD at 10 μM, or PEX at a concentration range (0.3 – 10 μM) were passed over the immobilized peptides for 6 min in Biacore buffer. Biacore sensorgrams showed specific binding measured in response units (RUs) for PEX (Panel A). The negative-control protein CBD did not bind P175L or P175S (Panel B). Plotting the binding of PEX to immobilized P175L and P175S peptides surfaces revealed that PEX had a concentration-dependent binding to P175L, but very weak binding to P175S (Panel C, D).
2.5. P175L but not P175S inhibits activation of proMMP-2
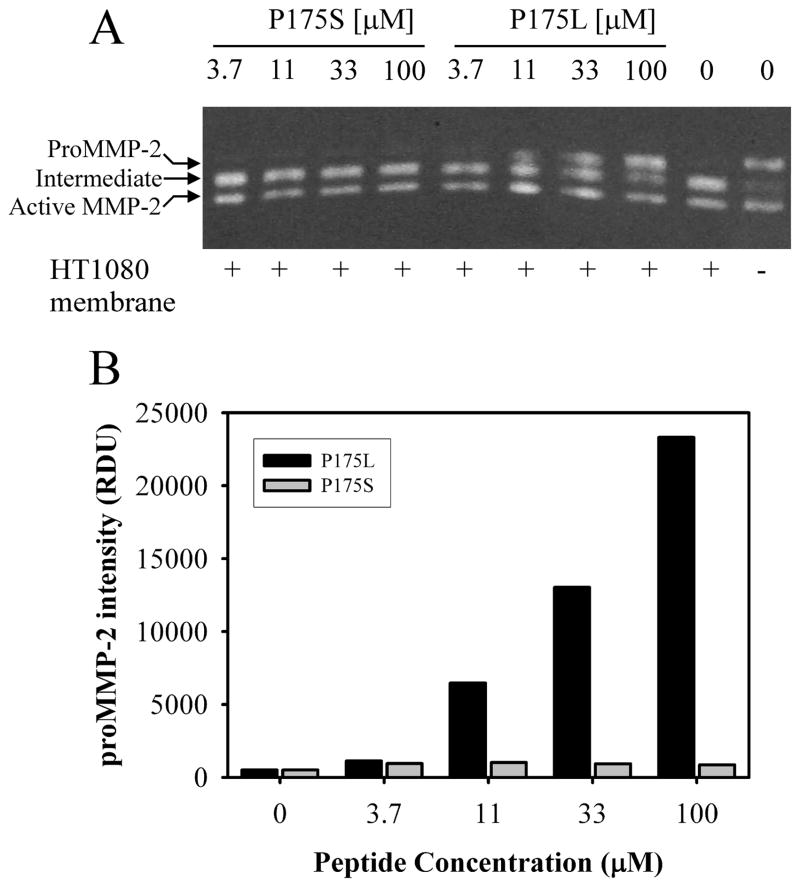
Having established that peptide P175L bound specifically to PEX, we measured whether this synthetic peptide could inhibit membrane-mediated activation of proMMP-2. When purified proMMP-2 was incubated with HT1080 membranes in the presence of a concentration range of P175L or P175S, P175L inhibited the activation of proMMP-2 in a concentration dependent manner (Fig. 5A, B). Thus, the amount of proMMP-2 was increased with increasing concentrations of P175L. In contrast, P175S, which bound very weakly to PEX, did not have any inhibitory effect on the cell membrane- mediated activation of proMMP-2.
Figure 5. P175L, but not P175S, inhibits activation of proMMP-2 by HT1080 cell membranes.
HT1080 cell membrane fraction was isolated and incubated with proMMP-2 alone (No inhibitor) or in the presence of a concentration range of P175L or P175S (3.7 – 100 μM). The reactions were analyzed by gelatin zymography (Panel A). Latent (66 kDa), intermediate (62 kDa) and active form of MMP-2 (59 kDa) were detected, and proMMP-2 was measured by densitometric analysis of digitized gels as a reflection of the inhibitory effects of the peptides on proMMP-2 activation. The analysis (Panel B) demonstrated that P175L could inhibit activation of proMMP-2 in a concentration dependent manner, whereas P175S had no effects on the activation. One representative experiment is shown and results were confirmed in repeated experiments.
Together, these experiments demonstrated clearly that isolated C-TIMP-2 as well as a synthetic binding site peptide from the tail of C-TIMP-2 can inhibit activation of proMMP-2.
3. Discussion
The N-terminal domain of TIMP-2 binds the catalytic site of MMP-2 to inhibit the activity of the enzyme, whereas the C-terminal domain of TIMP-2 binds the PEX domain of MMP-2 in the cellular activation complex (Howard and Banda, 1991). When TIMP-2 is bound to the PEX domain of MMP-2, the N-domain of TIMP-2 is oriented away from the MMP-2 active site (Morgunova et al., 2002). One molecule of TIMP-2 can not bind the catalytic site and the PEX domain of MMP-2 concurrently. TIMP-2 at higher concentrations inhibits activation of proMMP-2 (Strongin et al., 1993; Strongin et al., 1995) by two possible mechanisms: a). The N-terminal domain of TIMP-2 may bind directly to MT1-MMP and thereby inhibit the cleavage of the prodomain of proMMP-2 (Butler et al., 1998) or b) Excess of TIMP-2 occupies the binding site on PEX of free proMMP-2 and may thereby prevent the integration of proMMP-2 into the activation complex (Strongin et al., 1993). In fact, both isolated N-terminal domain of TIMP-2 and C-terminal domain of TIMP-2 inhibited proMMP-2 activation in cells (Kai et al., 2002; Butler et al., 1998). Our present results support the concept of two binding sites since C-TIMP-2 could bind specifically to PEX domain of MMP-2 (Fig. 1B) and fully compete the binding between PEX and TIMP-2, but not between MMP-2E404A and TIMP-2 (Fig. 3). This result indicated that our subsequent analyses of C-TIMP-2 effects on proMMP-2 activation were not confounded by possible effects of the N-terminal domain of TIMP-2. Complementing the observation that a C-TIMP-2 fusion protein at 4.5 μM could inhibit proMMP-2 activation in cells (Kai et al., 2002), we found that the inhibitory effects of C-TIMP-2 were concentration-dependent and inhibition of activation was observed at concentrations as low as 259 nM. Our study is consistent with results by others who found that full-length TIMP-2 inhibited activation of proMMP-2 at concentrations greater than 285 nM (Strongin et al., 1995). To extend previous kinetic analysis of the inhibition of MMP-2 by TIMP-2, which analyzed the interaction between C-TIMP-2 and PEX (Willenbrock et al., 1993; Overall et al., 2000), we found in protein – protein binding assays that C-TIMP-2 bound the PEX domain of MMP-2 with an apparent Kd of 1.7 × 10−7 M (Fig. 1B). This value is close to the reported Kdapp for the interactions between full-length TIMP-2 and PEX of 0.6 – 0.7 × 10−7 M (Bigg et al., 1997; Olson et al., 1997). Taken together, these analyses verified first that the interactions between the C-terminal domain of TIMP-2 and the PEX domain of proMMP-2 are important for activation of proMMP-2 and, secondly, demonstrated that successfully competing the binding between PEX and the C-terminal domain of TIMP-2 inhibited the activation of proMMP-2
In our quest to design peptide inhibitors of proMMP-2 activation, we reviewed available binding site information from studies of interactions between PEX and TIMP-2 (Fig 1D). The residues 568–631 on modules III and IV of PEX were previously found to be important for the binding to TIMP-2 in domain replacement (Butler et al., 1998), and results from subsequent single, double, and triple mutations of putative binding site residues pointed to TIMP-2 binding among several clusters of cationic residues in modules III and IV (Overall et al., 1999). The observations that non-contiguous positively charged residues on PEX were important for TIMP-2 binding, and that disruption of the tertiary structure of PEX by reduction of the disulfide bridges abrogated its capacity to inhibit proMMP-2 activation (Strongin et al., 1993) indicated that PEX did not contain a linear binding site motifs for TIMP-2 that could be mimicked in a synthetic peptide inhibitor. Indeed, several peptides derived from PEX synthesized by others did not inhibit proMMP-2 activation (Strongin et al., 1993). Therefore, we focused our efforts on TIMP-2 as a source of inhibitory peptides.
Crystal structure analysis of TIMP-2 in complex with MMP-2 identified a motif in the C-terminal tail of TIMP-2 that extended into a pocket-like structure on the surface of PEX (Morgunova et al., 2002). Moreover, it was shown that the TIMP-2 tail residues 187EFLDIEDP194 are essential for formation of a stable activation complex and that TIMP-2 without this tail did not support proMMP-2 activation (Worley et al., 2003). The observation that the acidic residues Glu192 and Asp193 in this tail segment are required for stability of the activation complex and activation of proMMP-2 (Kai et al., 2002; Worley et al., 2003) lent further support to continued analysis of inhibitory peptides from the tail region of TIMP-2.
Taking into consideration results from the above studies and the observation that a synthetic peptide containing only the last 13 residues of the TIMP-2 tail did not bind PEX in plate binding assays and that antibodies to this peptide region did not block the PEX – TIMP-2 interactions (Overall et al., 2000), we proceeded to design and test a 20 amino acid long synthetic peptide containing residues C175–P194 (P175L) and a shorter 11 amino acid control peptide (P175S, C175–K185). Analyses of these TIMP-2 tail derived synthetic peptides by SPR yielded results that were consistent with those from studies of truncated TIMP-2 (Worley et al., 2003). Specifically, P175L bound recombinant PEX in a concentration dependent manner and six fold stronger than peptide P175S (Fig. 4). The results match previous study that TIMP-2 with deleted residues 186–194 interact 5–7 fold slower with MMP-2 (Butler et al., 1998). In our proMMP-2 activation assays, P175L inhibited cell membrane-mediated activation of proMMP-2 whereas P175S had no such effect (Fig. 5). The fact that neither our 11-residue P175S nor a 13-residue TIMP-2 tail peptide (Overall et al., 2000) inhibited activation of proMMP-2 indicates that the N-terminal 7 residues as well as C-terminal 9 residues of P175L are critical for the binding between TIMP-2 and PEX of MMP-2 in the activation complex. Therefore we identified a specific peptide which inhibits cell membrane mediated activation of proMMP-2.
Augmenting its role in MMP inhibition, TIMP-2 has activities on cell growth promotion and inhibition, apoptosis, and inhibition of angiogenesis (Lambert et al., 2004). Those activities occur independently of MMP inhibition since reductively alkylated TIMP-2, devoid of MMP inhibitory activities, promotes cell growth (Hayakawa et al., 1994). The C-terminal domain of TIMP-2 expressed and purified with a yeast expression system reduces angiogenesis by inhibiting the proliferation of capillary endothelial cells (Fernandez et al., 2003). Further mapping of anti-angiogenic activities found that a 24-amino acid peptide corresponding to loop 6 of TIMP-2 is a potent inhibitor of angiogenesis (Fernandez et al., 2003). This region includes Met149 which is involved in forming the contact interface in the TIMP-2 and MMP-2 complex. It is noteworthy that a 19-amino acid tail peptide, which is only one residual shorter than P175L, had no anti-angiogenic activities (Fernandez et al., 2003). Thus, inhibition of angiogenesis and activation of proMMP-2 rely on different C-TIMP-2 motifs.
The successful use of peptides in phamocotherapy for inhibition of coagulation factors Via, IXa and Xa (Dennis et al., 2000; Izaguirre et al., 2009; Roberge et al., 2001), vasopressin (Manning et al., 2008), and caspase (Scheer et al., 2006), as well as promising applications of collagen-derived peptides for inhibition of MMP-1 (Lauer-Fields et al., 2009a), MMP-9 (Lauer-Fields et al., 2008), and MMP-13 (Lauer-Fields et al., 2009b) lend support and promise to the use of peptides with appropriate optimization for inhibitor development.
In summary, our experiments verified the critical contributions of the TIMP-2 tail residues to activation of proMMP-2 as was predicted in structural studies (Morgunova et al., 2002). Furthermore, the observation that a TIMP-2 tail derived synthetic peptide inhibited activation of proMMP2 by interrupting the binding between proMMP-2 and TIMP-2 confirmed the potential for designing a specific competitive peptide inhibitor of MMP-2 that act by inhibiting the activation of the enzyme.
4. Experimental procedures
4.1. Expression and purification of recombinant proteins
Recombinant human MMP-2E404A and collagen binding domain from MMP-2 (CBD) were expressed in E. coli and purified as described previously (Steffensen et al., 1995; Xu et al., 2004; Xu et al., 2007). The MMP-2E404A variant of MMP-2 has intact ligand binding properties, but no catalytic activities due to the active site Glu404Ala substitution (Morgunova et al., 2002). In addition, we here engineered new expression constructs for the hemopexin-like domain of MMP-2 (PEX), MMP-2E404A with deletion of PEX (MMP-2ΔPEXE404A), full-length TIMP-2 and the carboxyl-terminal domain of TIMP-2 (C-TIMP-2). All proteins were expressed with a His6 tag for affinity purification.
For the 42.4 kDa MMP-2E404AΔPEX, the coding region was amplified by PCR (forward primer, 5′-ccgctcgagTACAACTTCTTCCCTCGCAAG′3′; reverse primer, 5′-cggaattcTCACCCATAGAGCTCCTGAATGC-3′) from the pMMP-2E404A template (Xu et al., 2005a). Primers added the XhoI and EcoRI restriction sites (lower case) for directional cloning into the pRSETA expression vector (Invitrogen, San Diego, CA) as well as a stop codon. To obtain the 26.1 kDa PEX domain of MMP-2, we amplified the coding sequence from plasmid p186.2 (Collier et al., 1988) with forward primer, 5′-ctagctagcGGGGCCTCTCCTGACATTG-3′, and reverse primer, 5′-gggaagcttTCAGCAGCCTAGCCAGTCG-3′, and ligated the amplicon into NheI and HindIII restriction sites of the pGYMX expression vector (Steffensen et al., 1995). The coding sequences for the 26.3 kDa TIMP-2 and the 12.2 C-TIMP-2 were amplified from plasmid pGWIGH/htimp-2 (gift from Dr. Overall, University of British Columbia, Vancouver, Canada) using forward primer, 5′-ccgctcgagTGCAGCTGCTCCCCGGTG-3′, and reverse primer, 5′-cggaattcTTATGGGTCCTCGATGTCG-3′, for TIMP-2, and forward primer, 5′-ccgctcgagTGCAAGATCACGCGCTGC-3′, and reverse primer, 5′-cggaattcTTATGGGTCCTCGATGTCG-3′, for C-TIMP-2. Both amplicons were ligated into PRSETA using XhoI and EcoRI sites. All expression constructs were confirmed by double-stranded DNA sequencing at the Nucleic Acids Core Facility at the University of Texas Health Science Center at San Antonio (UTHSCSA).
The recombinant MMP-2ΔPEXE404A, PEX, TIMP-2 and C-TIMP-2 were expressed in E. coli BL21(DE3) as inclusion bodies. Proteins were solubilized with 8 M urea, 50 mM NaH2PO4, 300 mM NaCl, pH 8.0 and purified by Ni2+-affinity chromatography under denaturing conditions. Purified proteins were refolded by dialysis against phosphate buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) as detailed previously (Xu et al., 2004; Xu et al., 2005a; Xu et al., 2005b). The identities and masses of the recombinant proteins were verified by their predicted migration using SDS-PAGE and matrix-assisted laser desorption/ionization mass spectrometry (MALDI MS) at the UTHSCSA Institutional Mass Spectrometry Laboratory. All recombinant proteins were stored at −80°C until analysis.
4.2. Synthetic peptides
Two C-TIMP-2 tail peptides were synthesized (GenScript Corporation, Scotch plains, NJ). The 20 amino acid peptide P175L with the CAWYRGAAPPKQEFLDIEDP sequence corresponded to TIMP-2 tail residues 175–194. Peptide P175S corresponded to the 11 N-terminal residues of P175L and TIMP-2 residues 175–185 (CAWYRGAAPPK). Additional design rationale is presented in the Results section. No modifications were made at the N- and C- termini of the synthetic peptides. The synthetic peptides were in monomeric form as verified by MALDI-TOF under reducing and non-reducing conditions.
4.3. Protein-protein binding assays
To measure interactions of C-TIMP-2 with several proteins, 96-microwell plates were coated with recombinant MMP-2E404A, MMP-2ΔPEXE404A, PEX, or BSA as negative control at 0.5 μg/well in 0.1 M NaHCO3/Na2CO3, pH 9.6, overnight at 4 °C. Nonspecific binding sites were blocked with BSA (2.5%) for 1 h at 22 °C. The C-TIMP-2 was biotinylated and then equilibrated against 50 mM Tris, 150 mM NaCl, pH7.4 to remove residual biotin (Xu et al., 2005a). A concentration range (78 –1250 nM, and 0 nM) of biotinylated C-TIMP-2 was then reacted with the immobilized recombinant proteins or BSA for 1 h at 22 °C. After extensive rinses with 50 mM Tris, 150 mM NaCl, pH7.4/0.5% (v/v) Tween 20, bound C-TIMP-2 was detected with 1:10,000 diluted alkaline phosphatase-conjugated streptavidin (Pierce, Rockford, IL) and 1 mg/mL p-nitrophenyl phosphate disodium substrate (PNPP) (Sigma, St. Louis, MO), and quantified at 405 nm in an Opsys MR plate reader (Dynex, Chantilly, VA). All experiments were performed in duplicate and repeated at least twice. Apparent Kd values were calculated as previously (Steffensen et al., 2002) by nonlinear curve fitting from plots of C-TIMP-2 concentration versus bound protein (A405nm) using the equation y={(a–d)/[1+(x/c)b]} + d, where x is the concentration of C-TIMP-2 added, y is bound C-TIMP-2, b is the slope, c is the concentration of C-TIMP-2 at the inflection point (apparent Kd), a is minimum binding, and d is binding at saturation (Sigma Plot, SPSS Corp., Chicago, IL).
Competitive protein binding experiments tested the capacity of C-TIMP-2 to compete interactions of biotinylated MMP-2E404A or biotinylated PEX with coated full-length TIMP-2. Control experiments showed that unlabeled and labeled proteins had similar binding to coated ligands when bound proteins were detected using specific antibodies or AP-conjugated streptavidin. After coating plates with 0.5 μg/well TIMP-2, biotinylated proteins at concentrations yielding 50% of maximum binding, 50 nM for MMP-2E404A and 150 nM for PEX, were added to the coated wells alone (control) or simultaneously with a concentration range of competing C-TIMP-2 (10 – 5000 nM). The binding of the biotinylated proteins to TIMP-2 in the presence of C-TIMP-2 was measured with alkaline phosphatase-conjugated streptavidin and PNPP as described above and expressed as a function of the concentration of the competing C-TIMP-2. All experiments were performed in duplicate, and repeated at least twice.
4.4. Surface Plasmon resonance assays
Surface plasmon resonance (SPR) analyses of peptide-protein interactions were carried out at UTHSCSA Center for Surface Plasmon Resonance using a Biacore 3000 SPR instrument with CM5 sensor chips (GE Healthcare, Piscataway, NJ). The surfaces of the flow cells were immobilized with P175L or P175S. One uncoated reference cell served to adjust for non-specific binding. CM5 chips were activated by injection of 15 μl 0.2 M N-ethyl-N9-(dimethylaminopropyl) carbodiimide and 0.05 M N-hydroxy-succinimide for 6 min. Then the surfaces were modified by 2 - (2-pyridinyldithio) ethaneamine hydrochloride (PDEA Thiol Coupling Reagent, GE Healthcare). Peptides in acetate buffer, pH 4.0, were immobilized in the flow cells at 319 response units (RUs) for P175S and 327 RUs for P175L, followed by blocking with 30 μl cysteine (6 mg/ml). In peptide-protein binding assays, 30 μl recombinant PEX at concentrations from 0.3 – 10 μM in Biacore buffer (10 mM HEPES, 150 mM NaCl, 0.005% surfactant P20, pH 7.4) were passed over the coated surfaces. Purified collagen binding domain (CBD) of MMP-2 served as negative control (Xu et al., 2004). Interactions of proteins (analytes) with immobilized peptide ligand were expressed on RUs versus time sensorgrams. The surfaces were regenerated by injection of 6 M guanidine-HCl between analyses of peptide interactions with different proteins.
4.5. Inhibition of proMMP-2 activation by C-TIMP-2 in cell culture
Human fibrosarcoma HT1080 cells (ATCC, Rockville, Manasas, VA) were maintained in Dulbecco’s Modified Eagle Media (DMEM, Gibco, Rockville, MA) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. To measure the capacity of C-TIMP-2 to inhibit cellular activation of proMMP-2, 5 × 103 cells/well were seeded in tissue culture-treated 96-microwell plates and incubated for 3 days in the tissue culture incubator (37 °C, 5% CO2) to achieve confluence. Cells were rinsed with serum-free DMEM prior to addition of a concentration range of C-TIMP-2 (0.028 –7 μM, or 0 μM) in the presence of absence of 20 μg/ml concanavalin A (Con A) (Sigma, St. Louis, MO), which induces cellular activation of endogenous proMMP-2 (Overall et al., 2000). Conditioned media from the treated cells were collected after 24 h incubation and analyzed for latent and activated MMP-2 by zymography (See below). The experiments were repeated at least 3 times in duplicate, but results were only compared for experiments on the same plate.
4.6. Inhibition of proMMP-2 activation by C-TIMP-2 and synthetic peptides using isolated cell membrane
Activation of proMMP-2 by isolated HT1080 cell membranes was analyzed using established procedures (Knauper and Murphy, 2001) with slight modifications. In brief, HT1080 cells were cultured to confluence in DMEM with 10% FBS. After washes with PBS, the cells were incubated for 6 h with serum-free DMEM containing 10 μg/ml Con A. The cells were then scraped off the culture dishes mechanically into serum-free DMEM containing complete mini EDTA-free protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN) as recommended by the manufacturer. After sedimenting the cells at 2,500 × g for 5 min and washing two times with DMEM/protease inhibitor, cells were homogenized using a glass homogenizer in 20 mM Tris, pH 7.4, with protease inhibitors and passed through a 27G syringe needle 10 times. The homogenized cell suspensions were centrifuged at 28,000 × g for 10 min at 4 °C upon which the supernatant with the cell membrane fraction was further centrifuged at 100,000 × g for 90 min at 4 °C. The resulting cell membrane pellet was resuspended in 20 mM Tris, 10 mM CaCl2, 0.05% Brij35, pH 7.4, with protease inhibitor, quantified by the BCA assay (Pierce, Rockford, IL), and stored in 2 mg protein/ml aliquots at −80 °C until use.
In proMMP-2 activation assays, 50 pg proMMP-2 (Chemicon International Inc., Billerica, MA) and 0.5 μl of the HT1080 cell membrane preparation (1 μg) were incubated with concentration ranges of C-TIMP-2 (0.028 –7 μM, or 0 μM) or synthetic peptides P175L and P175S (3.7 – 100 μM, or 0 μM) in 10 μl reactions of 20 mM Tris, 10 mM CaCl2, 0.05% Brij35, pH 7.4, containing complete mini EDTA-free protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN) for 16 h at 22 °C. The levels of latent and activated MMP-2 were analyzed by gelatin zymography (See below). The experiments were repeated at least 3 times, comparing only experiments on the same plates.
4.7. Zymography
The presence of latent and activated forms of MMP-2 in conditioned cell culture media and proMMP-2 cell membrane activation assays was analyzed by zymography using 8% SDS-PAGE minislab gels co-polymerized with 150 μg/ml heat-denatured type I gelatin (BioRad, Hercules, CA) as detailed previously (Steffensen et al., 1995). Briefly, 10 μl of conditioned media or reactions were separated by electrophoresis under non-reducing conditions. The gelatin gels were then washed with 5% Triton X-100, incubated with 50 mM Tris, 200 mM NaCl, 5mM CaCl2, pH 7.2 overnight at 37 °C, and counterstained with Coomassie blue R-250. The activities of the 66 kDa pro, 62 kDa intermediate, and 59 kDa active forms of MMP-2, which appeared as translucent bands on a Coomassie Blue stained background, were captured in digitized images and band intensities were quantified within linear signal ranges using the Kodak 1D imaging software (Eastman Kodak, Rochester, NY).
Highlights.
Inhibition of activation of proMMP-2 by peptides was investigated.
Isolated C-terminal domain of TIMP-2 bound and inhibited activation of proMMP-2.
A 20-residue peptide from the tail of C-TIMP-2 inhibited activation of proMMP2.
The last 9 residues of C-TIMP-2 are essential for TIMP-2 interactions with proMMP-2.
Acknowledgments
We gratefully acknowledge Dr. Overall CM, Centre for Blood Research, University of British Columbia Vancouver, Canada for providing us with TIMP-2 plasmid and the Nucleic Acids Core Facility at the UTHSCSA for primers syntheses and DNA sequencing. We also thank Virgil Schirf at the UTHSCSA Center for Macromolecular Interactions and Dr. Susan Weintraub at the UTHSCSA Mass Spectrometry Laboratory for providing valuable help in the experiments. This work was supported by NIDCR grants DE018135, DE16312, DE17139, and DE014318. Work carried out in the institutional core laboratories was partially supported by NCI center grant P30 CA054174 to the San Antonio Cancer Center, and by funds from the UTHSCSA Executive Research Committee.
Abbreviations
- TIMP-2
tissue inhibitor of matrix metalloproteinase 2
- C-TIMP-2
carboxyl-terminal domain of TIMP-2
- ProMMP-2
latent matrix metalloproteinase-2
- MMP-2
matrix metalloproteinases-2
- MMP-2E404A
recombinant MMP-2 with active site Glu404 replaced by Ala
- PEX
hemopexin domain of MMP-2
- MMP-2ΔPEXE404A
recombinant MMP-2E404A with deleted PEX domain
- MT1-MMP
membrane type 1 matrix metalloproteinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong DG, Jude EB. The role of matrix metalloproteinases in wound healing. J Am Podiatr Med Assoc. 2002;92:12–18. doi: 10.7547/87507315-92-1-12. [DOI] [PubMed] [Google Scholar]
- Bigg HF, Shi YE, Liu YE, Steffensen B, Overall CM. Specific, high affinity binding of tissue inhibitor of metalloproteinases-4 (TIMP-4) to the COOH-terminal hemopexin-like domain of human gelatinase A. TIMP-4 binds progelatinase A and the COOH-terminal domain in a similar manner to TIMP-2. J Biol Chem. 1997;272:15496–15500. doi: 10.1074/jbc.272.24.15496. [DOI] [PubMed] [Google Scholar]
- Brown PD. Matrix metalloproteinase inhibitors. Breast Cancer Res Treat. 1998;52:125–136. doi: 10.1023/a:1006119319695. [DOI] [PubMed] [Google Scholar]
- Butler GS, Butler MJ, Atkinson SJ, Will H, Tamura T, van Westrum SS, Crabbe T, Clements J, d’Ortho MP, Murphy G. The TIMP2 membrane type 1 metalloproteinase “receptor” regulates the concentration and efficient activation of progelatinase A. A kinetic study. J Biol Chem. 1998;273:871–880. doi: 10.1074/jbc.273.2.871. [DOI] [PubMed] [Google Scholar]
- Butler GS, Overall CM. Updated biological roles for matrix metalloproteinases and new “intracellular” substrates revealed by degradomics. Biochemistry. 2009;48:10830–10845. doi: 10.1021/bi901656f. [DOI] [PubMed] [Google Scholar]
- Collier IE, Wilhelm SM, Eisen AZ, Marmer BL, Grant GA, Seltzer JL, Kronberger A, He CS, Bauer EA, Goldberg GI. H-ras oncogene-transformed human bronchial epithelial cells (TBE-1) secrete a single metalloprotease capable of degrading basement membrane collagen. J Biol Chem. 1988;263:6579–6587. [PubMed] [Google Scholar]
- Dennis MS, Eigenbrot C, Skelton NJ, Ultsch MH, Santell L, Dwyer MA, O’Connell MP, Lazarus RA. Peptide exosite inhibitors of factor VIIa as anticoagulants. Nature. 2000;404:465–470. doi: 10.1038/35006574. [DOI] [PubMed] [Google Scholar]
- Fernandez CA, Butterfield C, Jackson G, Moses MA. Structural and functional uncoupling of the enzymatic and angiogenic inhibitory activities of tissue inhibitor of metalloproteinase-2 (TIMP-2): loop 6 is a novel angiogenesis inhibitor. J Biol Chem. 2003;278:40989–40995. doi: 10.1074/jbc.M306176200. [DOI] [PubMed] [Google Scholar]
- Giannobile WV. Host-response therapeutics for periodontal diseases. J Periodontol. 2008;79:1592–1600. doi: 10.1902/jop.2008.080174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez DE, Alonso DF, Yoshiji H, Thorgeirsson UP. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur J Cell Biol. 1997;74:111–122. [PubMed] [Google Scholar]
- Hayakawa T, Yamashita K, Ohuchi E, Shinagawa A. Cell growth-promoting activity of tissue inhibitor of metalloproteinases-2 (TIMP-2) J Cell Sci. 1994;107 ( Pt 9):2373–2379. doi: 10.1242/jcs.107.9.2373. [DOI] [PubMed] [Google Scholar]
- Howard EW, Banda MJ. Binding of tissue inhibitor of metalloproteinases 2 to two distinct sites on human 72-kDa gelatinase. Identification of a stabilization site. J Biol Chem. 1991;266:17972–17977. [PubMed] [Google Scholar]
- Izaguirre G, Rezaie AR, Olson ST. Engineering functional antithrombin exosites in alpha1-proteinase inhibitor that specifically promote the inhibition of factor Xa and factor IXa. J Biol Chem. 2009;284:1550–1558. doi: 10.1074/jbc.M807340200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai HS, Butler GS, Morrison CJ, King AE, Pelman GR, Overall CM. Utilization of a novel recombinant myoglobin fusion protein expression system to characterize the tissue inhibitor of metalloproteinase (TIMP)-4 and TIMP-2 C-terminal domain and tails by mutagenesis. The importance of acidic residues in binding the MMP-2 hemopexin C-domain. J Biol Chem. 2002;277:48696–48707. doi: 10.1074/jbc.M209177200. [DOI] [PubMed] [Google Scholar]
- Knauper V, Murphy G. Methods for studying activation of matrix metalloproteinases. Methods Mol Biol. 2001;151:377–387. doi: 10.1385/1-59259-046-2:377. [DOI] [PubMed] [Google Scholar]
- Lambert E, Dasse E, Haye B, Petitfrere E. TIMPs as multifacial proteins. Crit Rev Oncol Hematol. 2004;49:187–198. doi: 10.1016/j.critrevonc.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Lauer-Fields JL, Chalmers MJ, Busby SA, Minond D, Griffin PR, Fields GB. Identification of specific hemopexin-like domain residues that facilitate matrix metalloproteinase collagenolytic activity. J Biol Chem. 2009a;284:24017–24024. doi: 10.1074/jbc.M109.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer-Fields JL, Minond D, Chase PS, Baillargeon PE, Saldanha SA, Stawikowska R, Hodder P, Fields GB. High throughput screening of potentially selective MMP-13 exosite inhibitors utilizing a triple-helical FRET substrate. Bioorg Med Chem. 2009b;17:990–1005. doi: 10.1016/j.bmc.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer-Fields JL, Whitehead JK, Li S, Hammer RP, Brew K, Fields GB. Selective modulation of matrix metalloproteinase 9 (MMP-9) functions via exosite inhibition. J Biol Chem. 2008;283:20087–20095. doi: 10.1074/jbc.M801438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning M, Stoev S, Chini B, Durroux T, Mouillac B, Guillon G. Peptide and non-peptide agonists and antagonists for the vasopressin and oxytocin V1a, V1b, V2 and OT receptors: research tools and potential therapeutic agents. Prog Brain Res. 2008;170:473–512. doi: 10.1016/S0079-6123(08)00437-8. [DOI] [PubMed] [Google Scholar]
- Morgunova E, Tuuttila A, Bergmann U, Tryggvason K. Structural insight into the complex formation of latent matrix metalloproteinase 2 with tissue inhibitor of metalloproteinase 2. Proc Natl Acad Sci U S A. 2002;99:7414–7419. doi: 10.1073/pnas.102185399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G, Houbrechts A, Cockett MI, Williamson RA, O’Shea M, Docherty AJ. The N-terminal domain of tissue inhibitor of metalloproteinases retains metalloproteinase inhibitory activity. Biochemistry. 1991;30:8097–8102. doi: 10.1021/bi00247a001. [DOI] [PubMed] [Google Scholar]
- Murphy G, Stanton H, Cowell S, Butler G, Knauper V, Atkinson S, Gavrilovic J. Mechanisms for pro matrix metalloproteinase activation. APMIS. 1999;107:38–44. doi: 10.1111/j.1699-0463.1999.tb01524.x. [DOI] [PubMed] [Google Scholar]
- Olson MW, Gervasi DC, Mobashery S, Fridman R. Kinetic analysis of the binding of human matrix metalloproteinase-2 and −9 to tissue inhibitor of metalloproteinase (TIMP)-1 and TIMP-2. J Biol Chem. 1997;272:29975–29983. doi: 10.1074/jbc.272.47.29975. [DOI] [PubMed] [Google Scholar]
- Overall CM, King AE, Sam DK, Ong AD, Lau TT, Wallon UM, DeClerck YA, Atherstone J. Identification of the tissue inhibitor of metalloproteinases-2 (TIMP-2) binding site on the hemopexin carboxyl domain of human gelatinase A by site-directed mutagenesis. The hierarchical role in binding TIMP-2 of the unique cationic clusters of hemopexin modules III and IV. J Biol Chem. 1999;274:4421–4429. doi: 10.1074/jbc.274.7.4421. [DOI] [PubMed] [Google Scholar]
- Overall CM, Lopez-Otin C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat Rev Cancer. 2002;2:657–672. doi: 10.1038/nrc884. [DOI] [PubMed] [Google Scholar]
- Overall CM, Tam E, McQuibban GA, Morrison C, Wallon UM, Bigg HF, King AE, Roberts CR. Domain interactions in the gelatinase A.TIMP-2. MT1-MMP activation complex The ectodomain of the 44-kDa form of membrane type-1 matrix metalloproteinase does not modulate gelatinase A activation. J Biol Chem. 2000;275:39497–39506. doi: 10.1074/jbc.M005932200. [DOI] [PubMed] [Google Scholar]
- Roberge M, Santell L, Dennis MS, Eigenbrot C, Dwyer MA, Lazarus RA. A novel exosite on coagulation factor VIIa and its molecular interactions with a new class of peptide inhibitors. Biochemistry. 2001;40:9522–9531. doi: 10.1021/bi010592d. [DOI] [PubMed] [Google Scholar]
- Rosenthal EL, Matrisian LM. Matrix metalloproteases in head and neck cancer. Head Neck. 2006;28:639–648. doi: 10.1002/hed.20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer JM, Romanowski MJ, Wells JA. A common allosteric site and mechanism in caspases. Proc Natl Acad Sci U S A. 2006;103:7595–7600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorsa T, Tjaderhane L, Konttinen YT, Lauhio A, Salo T, Lee HM, Golub LM, Brown DL, Mantyla P. Matrix metalloproteinases: contribution to pathogenesis, diagnosis and treatment of periodontal inflammation. Ann Med. 2006;38:306–321. doi: 10.1080/07853890600800103. [DOI] [PubMed] [Google Scholar]
- Stanley CM, Wang Y, Pal S, Klebe RJ, Harkless LB, Xu X, Chen Z, Steffensen B. Fibronectin fragmentation is a feature of periodontal disease sites and diabetic foot and leg wounds and modifies cell behavior. J Periodontol. 2008;79:861–875. doi: 10.1902/jop.2008.070492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffensen B, Wallon UM, Overall CM. Extracellular matrix binding properties of recombinant fibronectin type II-like modules of human 72-kDa gelatinase/type IV collagenase. High affinity binding to native type I collagen but not native type IV collagen. J Biol Chem. 1995;270:11555–11566. doi: 10.1074/jbc.270.19.11555. [DOI] [PubMed] [Google Scholar]
- Steffensen B, Xu X, Martin PA, Zardeneta G. Human fibronectin and MMP-2 collagen binding domains compete for collagen binding sites and modify cellular activation of MMP-2. Matrix Biol. 2002;21:399–414. doi: 10.1016/s0945-053x(02)00032-x. [DOI] [PubMed] [Google Scholar]
- Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J Biol Chem. 1995;270:5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- Strongin AY, Marmer BL, Grant GA, Goldberg GI. Plasma membrane-dependent activation of the 72-kDa type IV collagenase is prevented by complex formation with TIMP-2. J Biol Chem. 1993;268:14033–14039. [PubMed] [Google Scholar]
- Willenbrock F, Crabbe T, Slocombe PM, Sutton CW, Docherty AJ, Cockett MI, O’Shea M, Brocklehurst K, Phillips IR, Murphy G. The activity of the tissue inhibitors of metalloproteinases is regulated by C-terminal domain interactions: a kinetic analysis of the inhibition of gelatinase A. Biochemistry. 1993;32:4330–4337. doi: 10.1021/bi00067a023. [DOI] [PubMed] [Google Scholar]
- Williamson RA, Marston FA, Angal S, Koklitis P, Panico M, Morris HR, Carne AF, Smith BJ, Harris TJ, Freedman RB. Disulphide bond assignment in human tissue inhibitor of metalloproteinases (TIMP) Biochem J. 1990;268:267–274. doi: 10.1042/bj2680267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley JR, Thompkins PB, Lee MH, Hutton M, Soloway P, Edwards DR, Murphy G, Knauper V. Sequence motifs of tissue inhibitor of metalloproteinases 2 (TIMP-2) determining progelatinase A (proMMP-2) binding and activation by membrane-type metalloproteinase 1 (MT1-MMP) Biochem J. 2003;372:799–809. doi: 10.1042/BJ20021573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen Z, Wang Y, Bonewald L, Steffensen B. Inhibition of MMP-2 gelatinolysis by targeting exodomain-substrate interactions. Biochem J. 2007;406:147–155. doi: 10.1042/BJ20070591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen Z, Wang Y, Yamada Y, Steffensen B. Functional basis for the overlap in ligand interactions and substrate specificities of matrix metalloproteinases-9 and -2. Biochem J. 2005a;392:127–134. doi: 10.1042/BJ20050650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wang Y, Chen Z, Sternlicht MD, Hidalgo M, Steffensen B. Matrix metalloproteinase-2 contributes to cancer cell migration on collagen. Cancer Res. 2005b;65:130–136. [PubMed] [Google Scholar]
- Xu X, Wang Y, Lauer-Fields JL, Fields GB, Steffensen B. Contributions of the MMP-2 collagen binding domain to gelatin cleavage. Substrate binding via the collagen binding domain is required for hydrolysis of gelatin but not short peptides. Matrix Biol. 2004;23:171–181. doi: 10.1016/j.matbio.2004.05.002. [DOI] [PubMed] [Google Scholar]