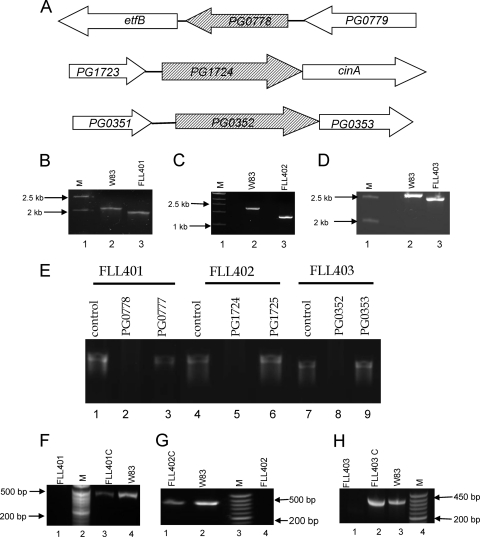
Fig. 2.
Construction and confirmation of P. gingivalis isogenic mutants. (A) Chromosomal structures of PG0778, PG1724, and PG0352. Hatched arrows indicate the coding genes, and white arrows indicate genes located downstream. The direction of the arrows indicates the transcription orientation. (B) Colony PCR confirmation of the mutant FLL401.Lane 1, molecular size markers; lane 2, PCR amplification of 2.2-kb fragment from W83; lane 3, 1.8-kb PCR amplicon. The primers used were 1 and 6 (see Table S2 in the supplemental material). (C) Colony PCR confirmation of the mutant FLL402. Lane 1, molecular size markers; lane 2. 1.7-kb PCR amplicon from W83; lane 3, 0.9-kb PCR amplicon from FLL402. The primers used were 7 and 12 (see Table S2 in the supplemental material). (D) Colony PCR confirmation of the mutant FLL403. Lane 1, molecular size markers; lane 2, 2.5-kb PCR amplicon from W83; lane 3, 2.3-kb PCR amplicon from FLL402. The primers used were 13 and 14 (see Table S2 in the supplemental material). (E) RT-PCR confirmation showing the nonpolar effects on the inactivated sialidase (PG0352) and sialoglycoproteases (PG0778 and PG1724) in isogenic defective mutants. Lane 1, PG0778 product using W83 cDNA as the template (472 bp); lane 2, PG0778 product using FLL401 cDNA as the template; lane 3, PG0777 product using FLL401 cDNA as the template (436 bp); lane 4, PG1724 product using W83 cDNA as the template (451 bp); lane 5, PG1724 product using FLL402 cDNA as the template; lane 6, PG1725 product using FLL402 cDNA as the template (447 bp); lane 7, PG0352 product using W83 cDNA as the template (360 bp); lane 8, PG0352 product using FLL403 cDNA as the template; lane 9, PG0353 product using FLL403 cDNA as the template (179 bp). (F) Transcription analysis of P. gingivalis mutant FLL401 by RT-PCR. The primers used were 19 and 20 (see Table S2 in the supplemental material). Lane 1, FLL401 mutant showing no amplification of the transcript; lane 2, molecular size markers; lane 3, complemented FLL401 showing a 472-bp amplified product; lane 4, wild-type W83. (G) Transcription analysis of P. gingivalis mutant FLL402 by RT-PCR. The primers used were 21 and 22 (see Table S2 in the supplemental material). Lane 1, complemented FLL401 showing 451-bp amplified product; lane 2, wild-type W83; lane 3, molecular size markers; lane 4, FLL401 mutant showing no amplification of the transcript. (H) Transcription analysis of P. gingivalis mutant FLL403 by RT-PCR. The primers used were 25 and 26 (see Table S2 in the supplemental material). Lane 1, FLL403 mutant showing no amplification of the transcript; lane 2, complemented FLL403 showing 360-bp amplified product; lane 3, wild-type W83; lane 4, molecular size markers.