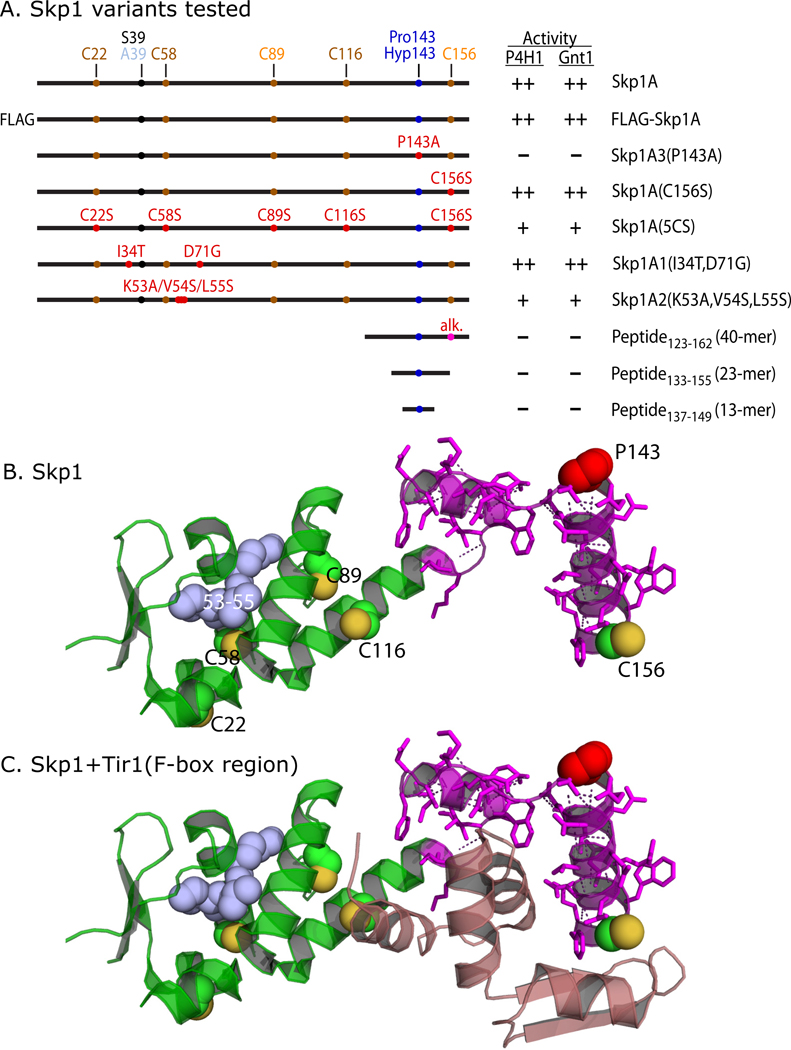
Figure 3. Summary of mutant constructs.
(A) Wild-type, mutant, and truncated Skp1 sequences are illustrated. Positions of the target Pro143, the S39A polymorphism which distinguishes Skp1A from Skp1B, the 5 Cys residues, and mutated amino acids, are indicated (see Fig. S4 for sequence). C89 and C156 are differentially colored to indicate their accessibility in the native protein. Corresponding substrates for Gnt1 were prepared by exhaustive hydroxylation by P4H1. Substrate activities of the Skp1 preparations toward P4H1 and Gnt1, from this study, are summarized: ++ = normal activity; + = low activity; − = no activity detected. (B) Crystal structure of Skp1 excerpted from a complex of Arabidopsis thaliana Ask1 (Skp1) with the TIR1 F-box protein and auxin (32; PDB 2P1N), using PyMol. Pro143 is in red; S-atoms of Cys residues (substituted according to Dictyostelium sequence based on the alignment in Fig. S4) are in yellow; the 40-mer peptide (with polar contacts shown) is in magenta; the consecutive point mutations of Skp1A2 are in blue. (C) Amino acids 8–74 of TIR1, which includes its F-box domain, is included in mauve.