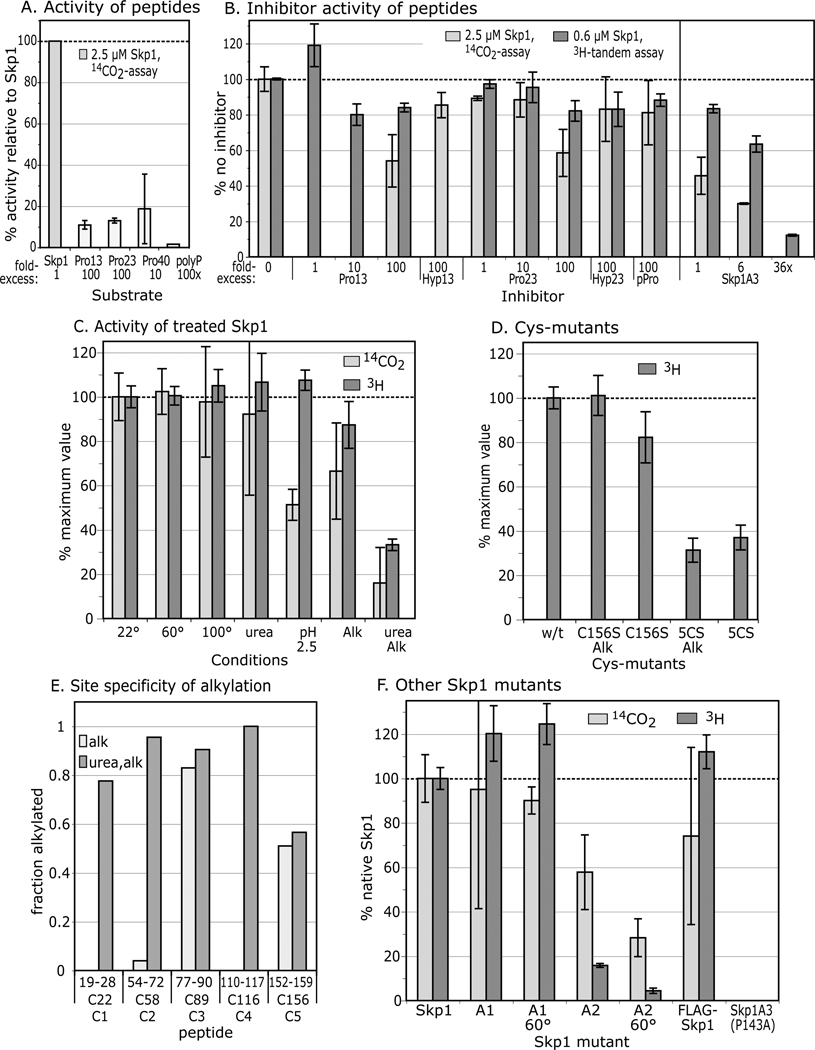
Figure 4. Substrate activity of Skp1 peptides, and denatured, alkylated, and mutant Skp1s.
(A) Comparison of Skp1 peptides and poly-L-Pro with Skp1A, provided at the concentrations indicated, using the 14C-release assay. Average values from 2–3 independent trials, ±SEM, are shown. (B) Effects of peptides and mutant Skp1A3(P143A) on the reaction with Skp1A, using either the 14C-release assay (Skp1A = 5 µM) or the coupled, 2-step 3H-assay (Skp1 = 0.63 µM). (C) Skp1A was subjected to denaturing conditions and/or alkylation with iodoacetamide, and restored to native reaction buffer as required. Recovery was verified by SDS-PAGE (Fig. S1). (D) The all 5Cys-Ser mutant Skp1 (5CS) and Skp1A(C156S) were untreated or alkylated (without urea). Data are presented as average values ± SEM (100% = 12,000–36,000 3H dpm; 8000–16,000 14C dpm). (E) Mapping of alkylation sites. After iodoacetamide treatment in the presence or absence of 6 M urea, Skp1 samples were digested with endo Lys-C and analyzed by MALDI-TOF-MS. Ion currents corresponding to alkylated Cys-peptides, relative to total ions associated with Cys-peptides (non-alkylated and disulfides) are reported. (F) Skp1A1, Skp1A2, FLAG-Skp1 and Skp1A3(P143A) were prepared from E. coli under non-denaturing conditions, and assayed at 2.5 µM using the 14C-release assay (100% activity = 24,600 dpm), or at 0.63 µM using the coupled 3H-GlcNAc two-step assay, at 22 ° or 60 °C as indicated.