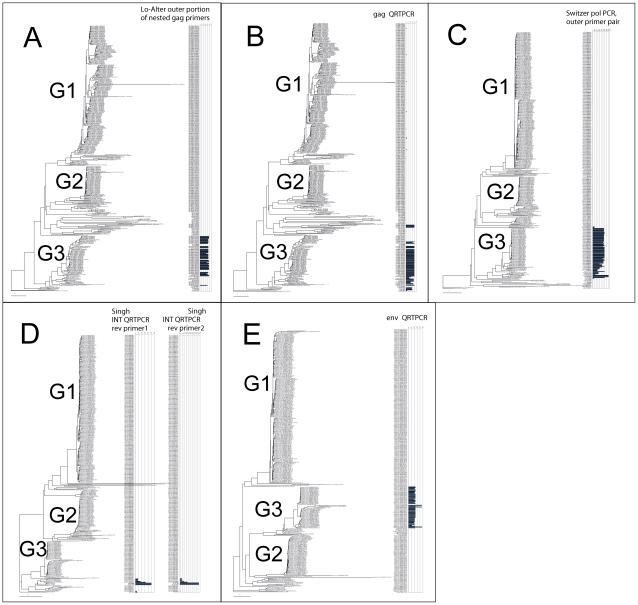
Figure 3. Estimated PCR detection range in an alignment of 300 high-scoring gammaretroviruslike proviruses found by RetroTector in the mm8 assembly.
Predictions were mapped onto NJ trees of alignments of gag, pol, integrase and env nucleotide sequences. A PCR detection score (shown as horizontal bars) was calculated by multiplying the fit of primers and probe (if present) to the target sequences for each provirus in the alignment. The degree of fit (match) was estimated using a modified NucZip algorithm [28]. A more complete treatment of this subject will be published separately (Danielsson et al, in preparation). The trees and alignments are further presented, and shown in higher resolution together with trees made with several algorithms, with bootstrap figures, in Information S1. A. A gag nt alignment assessed with outer primers of the nested Lo/Alter PCR [3] (HMRV sequences from that paper are shown in red), and B. the same tree with the prediction for the primers and probe of the gag RTQPCR presented in this paper. C. A pol tree, with the prediction for the outer primer pair of the nested RT-based PCR of Switzer et al [5] is shown. D. A tree based on the 3′ (mainly integrase) portion of pol. Predictions based on the Singh RTQPCR, with its two reverse primers [35], are shown. XMRV sequences are shown in red. E. An env tree. The prediction of the detection range of the env RTQPCR presented in this paper is shown. A higher resolution picture is shown in Information S1.